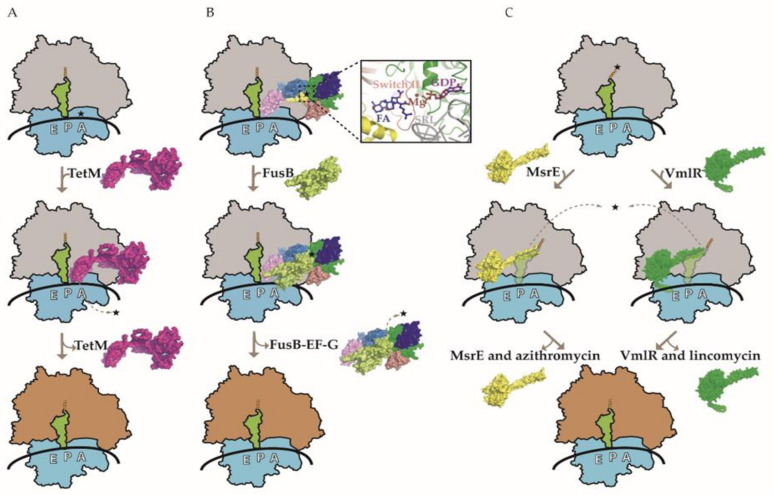
Figure 1.
Three models of ribosome protection protein-mediated antibiotic resistance. (A) A model for ribosome protection against tetracycline (TET) mediated by the TetM protein. Drug-stalled ribosome with tRNA in the P-site (green) is rescued by TetM (pink), which competes with TET (shown with star) in the A-site, thereby purging it from the ribosome. The subsequent GTP hydrolysis-dependent release of TetM from the ribosome enables protein synthesis to resume. (B) A model for ribosome protection against fusidic acid (FA) mediated by the FusB protein. FA interaction with elongation factor G (EF-G) prevents its dissociation from the ribosome. FusB (lime green) interacts with the ribosome-bound EF-G, leading to its release and allowing translation to proceed in the presence of FA. The domains G, G’, II, III, IV, and V of EF-G are colored green, blue, deep salmon, yellow, and sky blue, respectively. An enlarged view of the FA binding pocket is shown, involving EF-G domains G, II, and III. EF-G switch II (residues 82–102) is colored red and the 23S ribosomal RNA sarcin-ricin loop (SRL) is colored gray. In addition, GDP and Mg2+ in the vicinity of FA are also shown. Notably, FusB does not interact with the same region of EF-G as FA and there is no evidence for direct physical displacement of the drug. (C) A model for ribosomes protection against various classes of PTC/NPET-targeting AB mediated by the ARE ABC-F proteins. Two representatives of ARE ABC-F proteins, MsrE (yellow) and VmlR (dark green), are shown to bind to the E-site of the drug-stalled ribosome. Their antibiotic resistance domain (ARD) distorts the tRNA in the P-site (green) in order to access the drug-binding site. Allosteric and/or steric interactions in PTC promote the dissociation of drugs. ATP binding may promote RPP–ribosome interaction, while ATP hydrolysis leads to the dissociation of RPP from the ribosome. Drugs corresponding to MsrE and VmlR are azithromycin and lincomycin, respectively (also shown with stars). Ribosomes with gray and orange large subunits represent translationally inactive and active complexes, respectively.