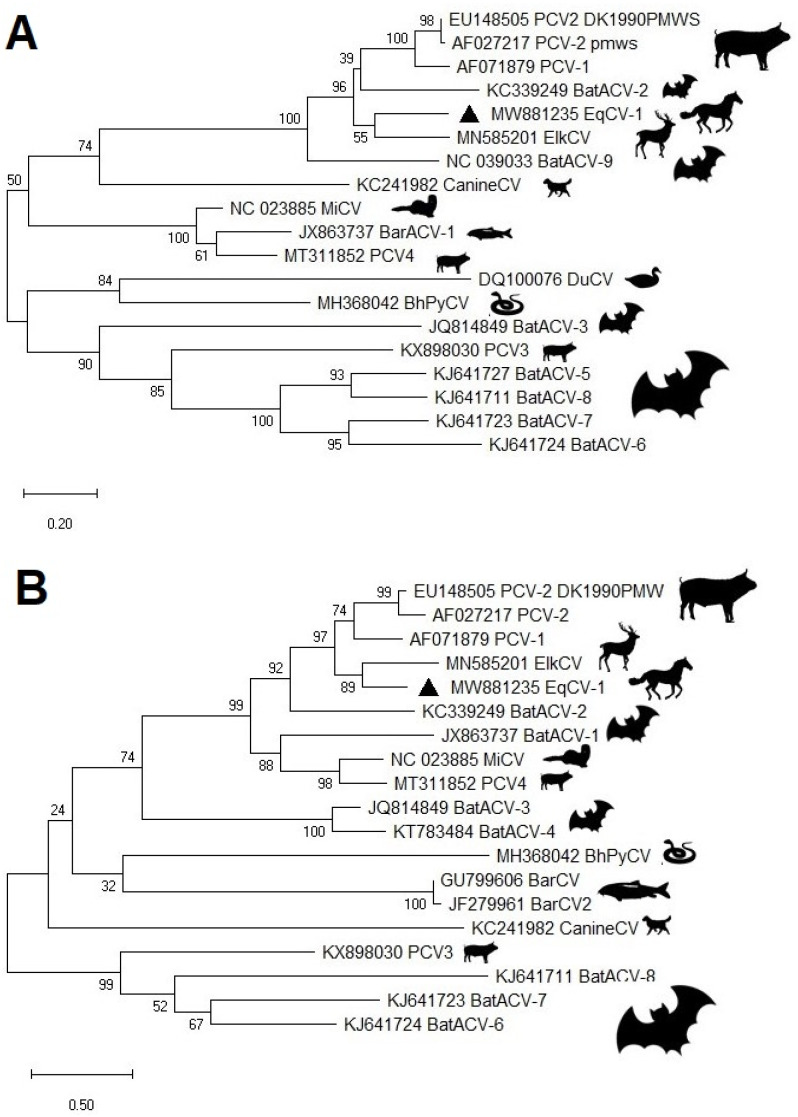
Figure 2.
(A). Phylogenetic analysis of EqCV1-Charaf replicase. (B). Phylogenetic analysis of EqCV1-Charaf capsid. The scale indicates amino acid substitutions per position. The amino acid (aa) pairwise alignments were performed with Genious software using the in-built MAFFT algorithm. The phylogenetic trees were constructed using the Maximum likelihood method with substitution model: Le Gascuel 2008 based model with gamma-distributed (G+) for Rep and Cap in MEGA software version X [33,41].