Figure 4.
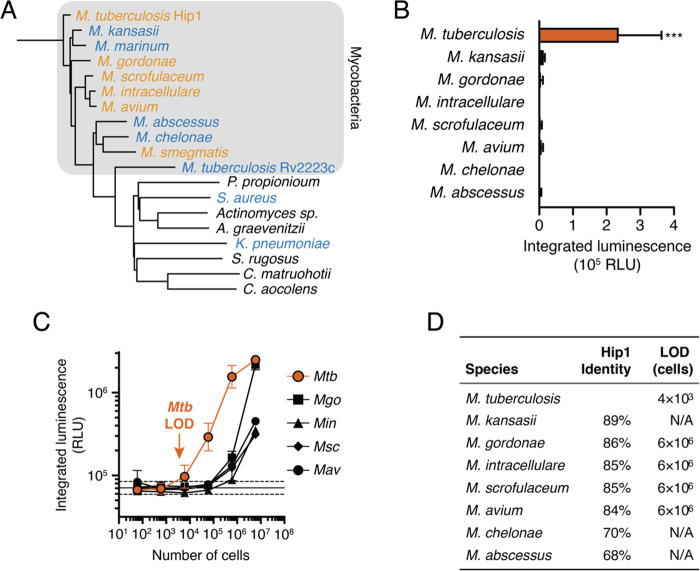
FLASH is selective for Mtb. (A) Phylogenetic tree of potential Mtb Hip1 homologues found in NTMs, other lung pathogens, and commensal members of the human airway and oral microbiomes. Also included is Rv2223c, an uncharacterized peptidase encoded by Mtb with sequence similarity to Hip1. Bacteria are colored by their ability to process FLASH at high cell densities (orange, active; blue, inactive; black, not tested). (B) FLASH signal for 6 × 104 cells of each NTM in 7H9 medium (mean ± SD, n = 3). Each sample was compared to the no-bacteria control via one-way ANOVA with Dunnett’s test (***, p < 0.001). (C) FLASH signal for M. tuberculosis (Mtb), M. gordonae (Mgo), M. intracellulare (Min), M. scrofulaceum (Msc), and M. avium (Mav) at the indicated cell number (mean ± SD, n = 3). (D) Sequence similarity of potential homologues to Mtb Hip1 and limits of detection calculated from part C for each NTM.