Fig. 6 |. Single-cell omics in direct reprogramming.
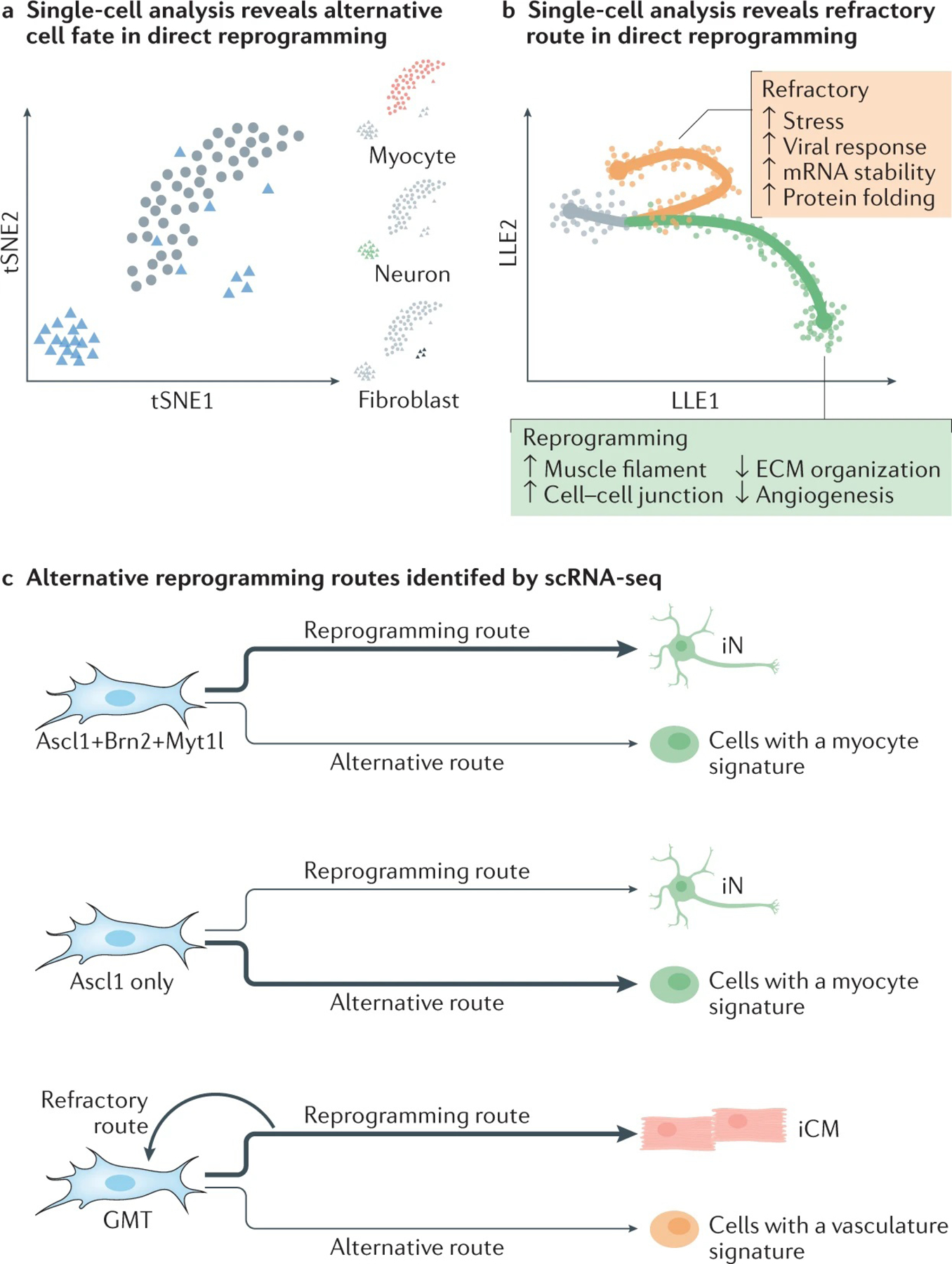
Computational approaches that produce information on reprogramming trajectories based on single-cell RNA-seq (scRNA-seq) data facilitate the identification of alternative routes in both direct neuronal and cardiac reprogramming. Two examples of the type of information that can be obtained are shown. a | Clustering analysis based on scRNA-seq at the late stages of Ascl1-mediated (circle) or BAM-mediated (triangle) neuronal reprogramming showed three distinct cell clusters with specific lineage gene expression of neuron (red), fibroblast (blue) and myocyte (green) fate, suggesting the existence of an alternative cell fate at the late stage of neuronal reprogramming. The plot is modified based on the data from REF.77. b | Trajectory analysis revealed two separate paths in human cardiac reprogramming. When cells engage in a ‘reprogramming path’, they gradually acquire a cardiomyocyte cell fate; however, cells can also follow a ‘refractory route’ and revert towards a fibroblast cell fate. Differential gene expression analysis identified genes involved in different pathways or cellular processes that are activated or suppressed while cells follow either route. The plot is modified based on the data in REF.76. c | The alternative reprogramming outcomes of direct reprogramming revealed by scRNA-seq analysis. In neuronal reprogramming mediated by Ascl1 only, most of the cells gained a transcription programme similar to myocytes. The addition of Brn2 and Myt1l suppressed the aberrant myogenic programme. In cardiac reprogramming mediated by Gata4, Mef2c and Tbx5 (GMT), most of the cells successfully gained a cardiac programme as expected. A small population of cells gained transcription signatures of vasculature and blood vessel development. scRNA-seq analysis of human cardiac reprogramming also revealed the existence of a refractory route where the cells reverted to a fibroblast fate. ECM, extracellular matrix; iCM, induced cardiomyocyte; iN, induced neuron; LLE, locally linear embedding; tSNE, t-distributed stochastic neighbour embedding. Part a adapted from REF.76, Springer Nature Limited; part b adapted with permission from REF.77, Elsevier.
