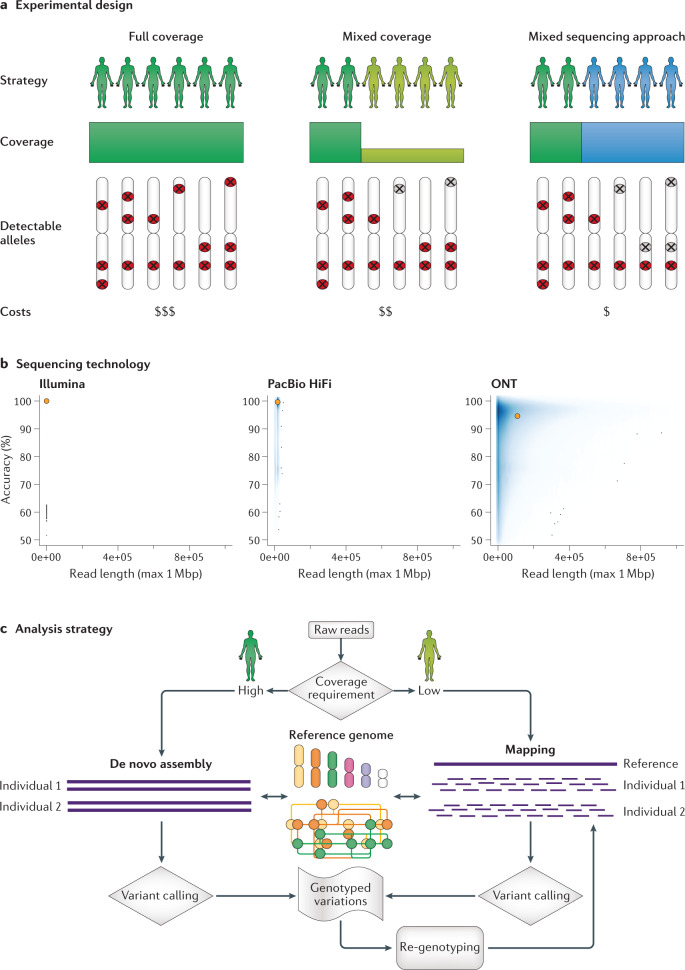
Fig. 2. Overview of long-read population study design.
a | The experimental design of three different approaches is outlined. In the first strategy (left), all samples are sequenced at medium to high coverage by long-read sequencing. In the second approach (middle), a proportion of the samples are sequenced with medium to high coverage and the remainder using low coverage by long-read sequencing (similar to the initial 1000 Genomes project). In the third approach (right), a proportion of the samples are sequenced at medium to high coverage by long-read sequencing and the remainder by short-read sequencing. The decision of which approach to take will affect the ability to detect common (red symbols) or rare (grey symbols) events in the population. The decision also depends on the available budget, existing data and the sample DNA availability. b | Overview of current established sequencing technologies based on CHM13 sequencing data79: Illumina, Pacific Biosciences (PacBio) High Fidelity (HiFi) reads or ultra-long reads from Oxford Nanopore Technologies (ONT). The N50 read length and average read accuracy are highlighted in orange. Although each technology has advantages and disadvantages, HiFi and ONT are the most promising for future applications. c | Overview of analysis strategies. Although multiple approaches are available, the main decision is whether to use an alignment-based approach or a de novo assembly-based approach, which has implications for sequencing requirements and the approaches, resolution and comprehensiveness of downstream computational analysis.