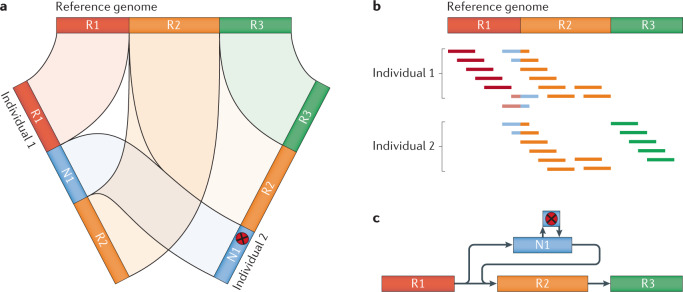
Fig. 3. Potential problems for different genome comparison approaches.
a | Schematic depiction of a potential problem in a de novo assembly-based approach. The presence of a novel segment N1 in two de novo assemblies, at different locations and, even more so, a sequence variant (red x), poses a challenge to correct reporting by current state-of-the-art methods and variation formats. b | Similar representation of the N1 problem in an alignment-based approach, where the coordinates of N1 are shared, but remain challenging for the identification of the single-nucleotide variant (SNV) or the entire N1 sequence. c | A graph-based representation of N1, which enables a clearer comparison of the variant across the samples, illustrating the potential benefits of graph genomes. R1–R3 represents the backbone of the graph genome and N1, and its SNV represents novel sequencing for a given sample set.