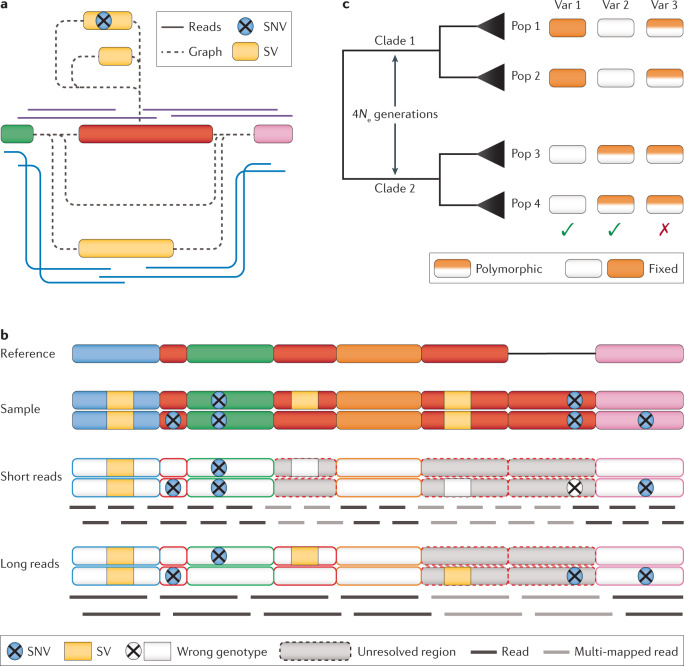
Fig. 4. Genotyping of SVs and SNVs across a population set.
a | Graph genome-based genotyping of a region with multiple alleles between two genome segments (green and pink). Insertions of different sizes (yellow) can be genotyped at the same locus using spanning reads (blue and purple) to identify the presence of two different alleles. b | An example of structural variants (SVs) and single-nucleotide variants (SNVs) across different unique and repeat regions being correctly or incorrectly genotyped based on read length. c | A phylogenetically informed filtering approach for SVs. Assuming that after a sufficiently long time (4Ne generations, where e = effective population size) most or all genetic variation should be fully sorted between two clades; variants that do not adhere to this assumption and are polymorphic across clades (for example, variant 3) can be removed. Although this approach is certainly very conservative and ignores the fact that some types of variation exhibit repeated mutations on the same locus, it can be considered a first step towards more reliable genotyping of SVs.