Figure 1. Genomic approaches for studying drug resistance.
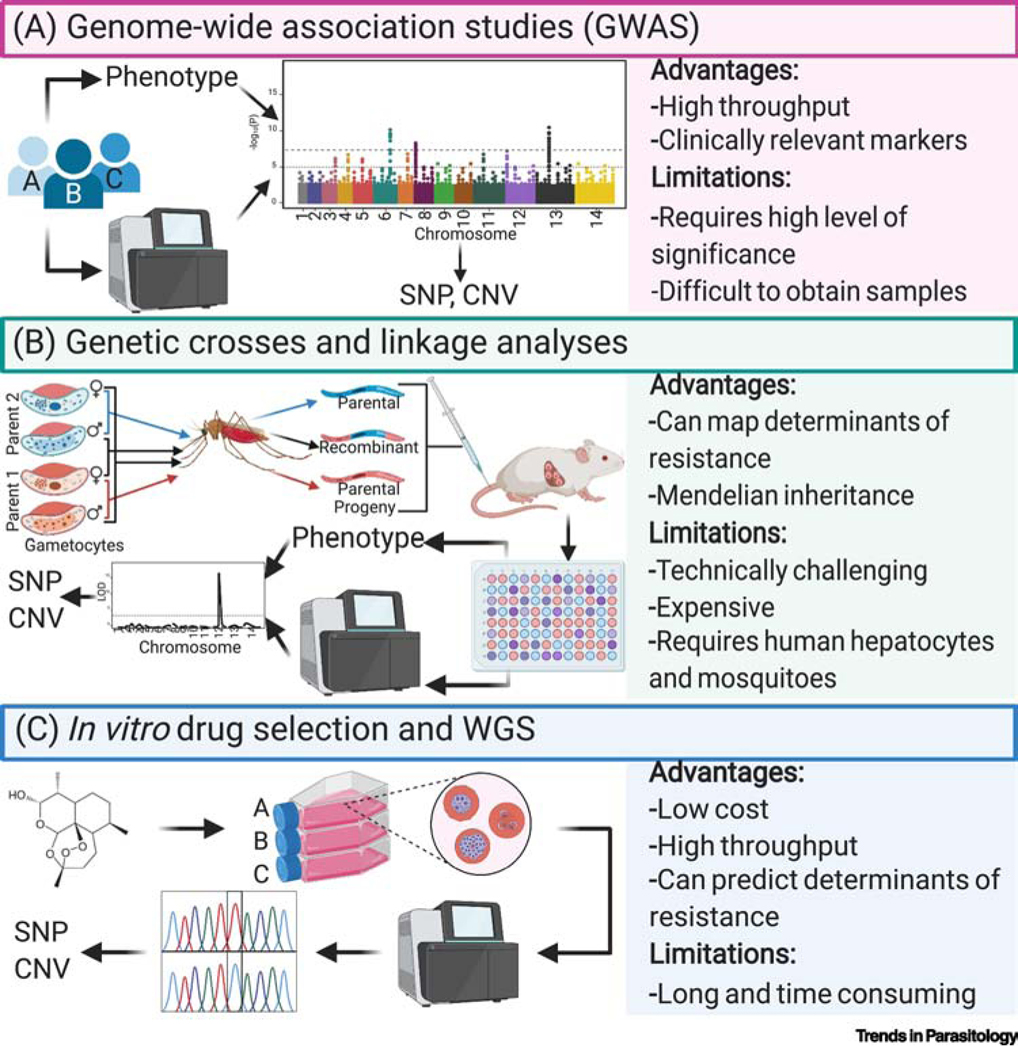
(A) Genome-wide association studies (GWAS): P. falciparum parasites are isolated from infected patients, profiled in vitro against antimalarial drugs of interest, and genotyped (using WGS, microsatellite markers or oligonucleotide arrays). These datasets are analyzed (as depicted visually in the Manhattan plot) to identify genetic variants associated with resistance. (B) Genetic crosses and linkage analyses: gametocytes from genetically distinct parental parasites are fed to Anopheles mosquitoes wherein they undergo sexual reproduction and genetic recombination, creating both parental and new recombinant genotypes. The resulting sporozoite haploid progeny are inoculated into FRG huHep mice by intravenous injection or mosquito bite, after which they develop inside the engrafted human hepatocytes and complete the liver to blood-stage transition after injection of human RBCs 6 to 7 days later. Progeny are cloned by limiting dilution and then genotyped and phenotyped. QTL analysis is conducted (as shown in the LOD plot) to localize the genetic determinant(s) of resistance and the polymorphisms that associate with the drug response variation. (C) in vitro drug selection and WGS (also known as in vitro evolution and whole-genome analysis [114]): antiplasmodial compounds are used to pressure P. falciparum parasites to select for recrudescent, resistant parasites. Genomic DNA (gDNA) of bulk cultures or their clones is sent for WGS, along with the parental strain, to identify genetic polymorphisms such as SNPs and CNVs that may underlie the resistance phenotype. WGS: whole-genome sequencing; SNP: single nucleotide polymorphism; CNV: copy number variation.
