Optimizations of reaction conditionsa.
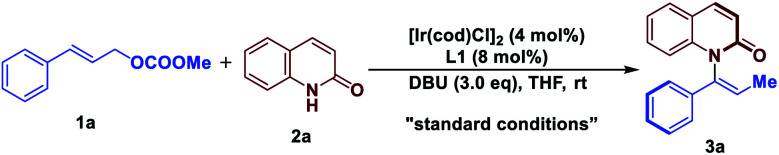
| |||
|---|---|---|---|
| Entry | Variation from standard conditions | eeb (%) | Yieldc (%) |
| 1 | None | 92 | 88 |
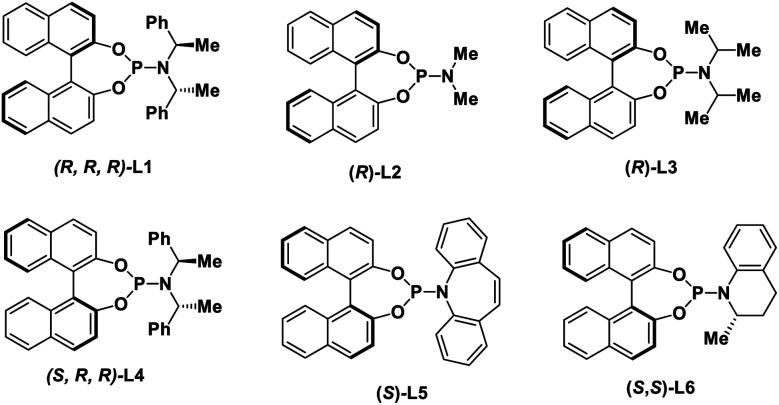
| 2 | L2 instead of L1 as ligand | — | Trace |
| 3 | L3 instead of L1 as ligand | 13 | <5% |
| 4 | L4 instead of L1 as ligand | — | Trace |
| 5 | L5 instead of L1 as ligand | −80 | — |
| 6 | L6 instead of L1 as ligand | 72 | 33 |
| 7 | Et3N instead of DBU as base | 89 | 33 |
| 8 | DABCO instead of DBU as base | 88 | 35 |
| 9 | TBD instead of DBU as base | 86 | 37 |
| 10 | 1,4-dioxane instead of THF as solvent | 91 | 86 |
| 11 | Toluene instead of THF as solvent | 87 | 64 |
| 12 | –OBz instead of –OCOOMe | 93 | 38 |
| 13 | –Cl instead of –OCOOMe | –3 | 46 |
| 14 | –OPO(OEt)2 instead of –OCOOMe | 50 | 42 |
| 15 | –OBoc instead of –OCOOMe | 89 | 34 |
| |||
Reaction conditions: all reactions were run on 0.1 mmol scale with respect to 1.
ee determined by chiral HPLC.
Isolated yield. DABCO = 1,4-diazabicyclo[2.2.2]octane, DBU = 1,8-diazabicyclo[5.4.0]undec-7-ene, TBD = 1,5,7-triazabicyclo[4.4.0]dec-5-ene.
