Figure 4.
In bovine zygotes, centrosome positions determine the sites of chromosome clustering and accuracy of chromosome segregation
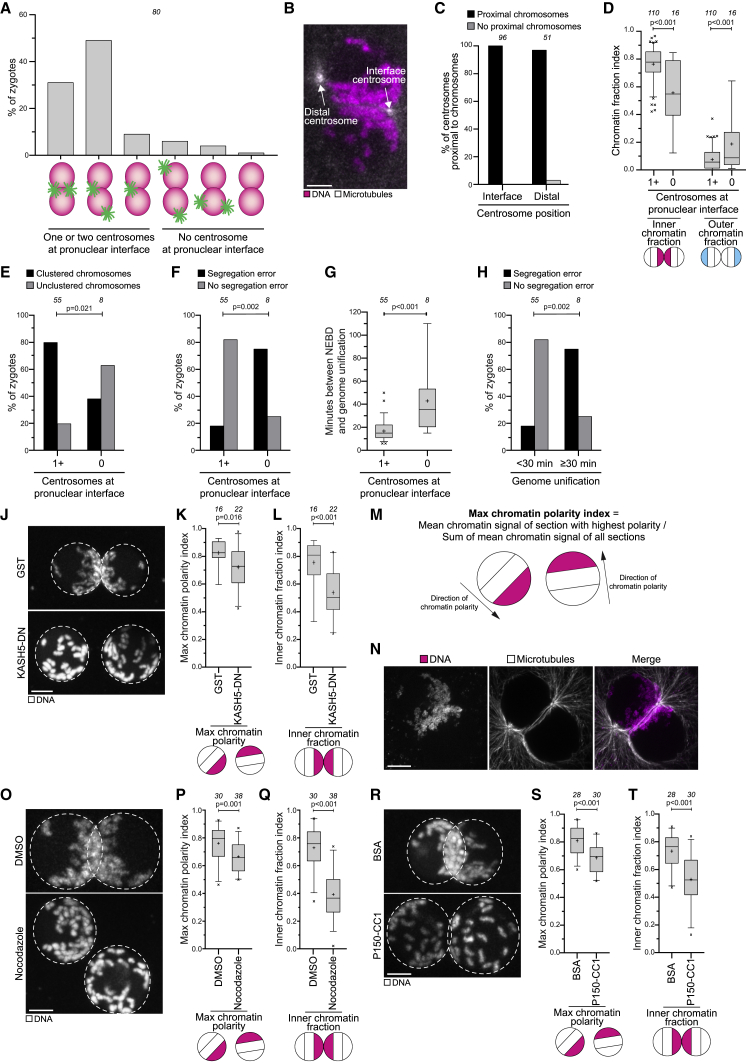
(A) Centrosome localization before NEBD in bovine zygotes.
(B) Representative still from a time-lapse movie of a zygote before NEBD. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Arrows specify distal and the interface centrosomes.
(C) Distal and interface centrosomes as in (B) were scored for proximity to chromosomes.
(D) Inner and outer chromatin fraction indices in zygotes with or without centrosomes at the pronuclear interface.
(E) Chromosome clustering was scored in zygotes with or without centrosomes at the pronuclear interface.
(F) Zygotes with or without centrosomes at the pronuclear interface were scored for the presence of chromosome segregation errors.
(G) Time between NEBD and the unification of the parental genomes in zygotes with or without centrosomes at the pronuclear interface.
(H) Zygotes in which parental genome unification took place within 30 min after NEBD or later were scored for the presence of chromosome segregation errors.
(J) Representative images of zygotes expressing GST or KASH5-DN. White, DNA (H2B-mScarlet). Dashed lines mark pronuclei. Z projections, 9 sections every 3.08 μm.
(K and L) Max chromatin polarity (see M) and inner chromatin fraction indices in zygotes expressing GST or KASH5-DN.
(M) Schematic illustrating calculation of the max chromatin polarity index.
(N) Representative immunofluorescence images of a zygote with microtubules wrapped around the pronuclei. White, microtubules (α-tubulin). Magenta, DNA (DAPI). Z projection, 35 sections every 0.1 μm.
(O–T) Representative images, max chromatin polarity and inner chromatin fraction indices in zygotes treated with DMSO or nocodazole before pronuclear juxtaposition (O–Q) or injected with BSA or P150-CC1 (R–T). The last time point before NEBD is shown. White, DNA (H2B-mScarlet). Dashed lines mark pronuclei. Z projections, 8 (O) and 12 (R) sections every 2.50 μm.
Zygotes having a pronucleus with uncondensed chromatin at NEBD were excluded from the quantifications in (C)–(H), (K), (L), (S), (T), (P), and (Q) to avoid accounting for the role of incomplete chromosome condensation at NEBD. Data are from eleven (A–H), four (K, L, S, and T), or six (P and Q) independent experiments. The number of analyzed zygotes (A and E–H), centrosomes (C), and pronuclei (D, K, L, S, T, P, and Q) is specified in italics. p values were calculated using unpaired two-tailed Student’s t test (D, G, K, L, S, T, P, and Q) and Fisher’s exact test (E, F, and H). Scale bar, 10 μm.
See also Figures S5 and S7 and Video S5.