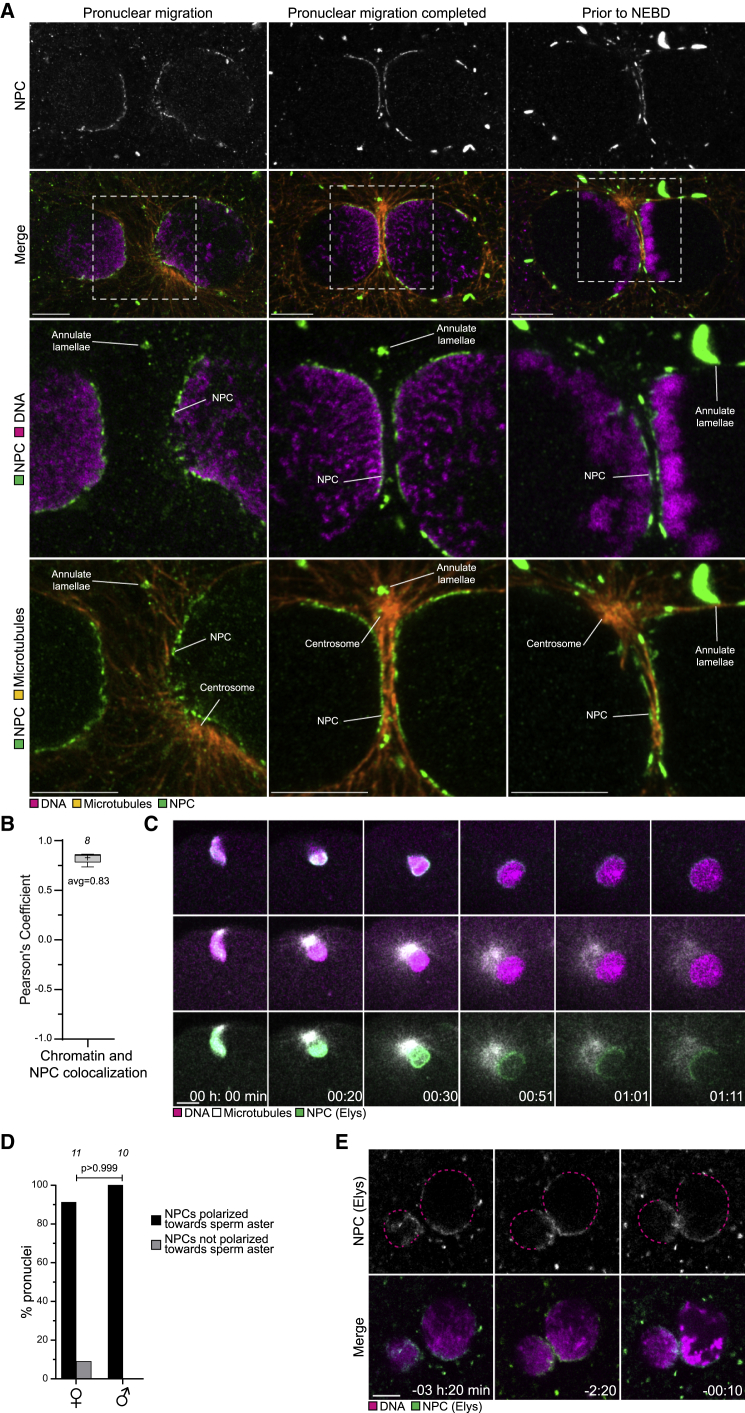
Figure 5.
Nuclear pore complexes cluster with chromatin at the pronuclear interface in bovine zygotes
(A) Representative immunofluorescence images of bovine zygotes during pronuclear migration (left), in early stages on pronuclear juxtaposition when chromatin condensation is incomplete (middle), and in late stages after increased chromatin condensation (right). Orange, microtubules (respectively, β-tubulin, α-tubulin, and α-tubulin). Magenta, DNA (DAPI). Green, NPC (respectively Nup98, NPC-Mab414, and NPC-Mab414). Outlined regions magnified in the bottom two rows. Centrosomes, NPCs, and annulate lamellae are indicated. Single sections Airyscan microscopy.
(B) Pearson’s coefficient quantifying the co-localization of NPCs and chromatin at the nuclear envelope. +1 indicates perfect co-localization, −1 indicates exclusion.
(C) Representative stills from time-lapse movies of zygotes expressing bElys-mClover3 (NPC, green), mScarlet-MAP4-MTBD (microtubules, white), and H2B-miRFP (DNA, magenta). 00:00 is start of image acquisition. First two time points are single confocal microscopy sections, whereas the other time points are Z projections of 3 (00:30) or 5 (00:51 to 01:11) sections every 2.50 μm.
(D) Female (♀) and male (♂) pronuclei were scored for polarization toward the sperm aster.
(E) Representative stills from time-lapse movies of zygotes expressing bElys-mClover3 (NPC, green) and H2B-mScarlet (DNA, magenta). The right pronucleus re-orients, leading to a more clustered organization of chromatin and NPCs. Dashed line indicates regions of the nuclear envelope devoid of NPCs. Time, h:min, 00:00 is NEBD. Single confocal microscopy sections.
Data are from two (B) or three (D) independent experiments. The number of analyzed zygotes (C) or pronuclei (D) is specified in italics. p value in (D) was calculated using unpaired two-tailed Student’s t test. Scale bars, 10 μm.