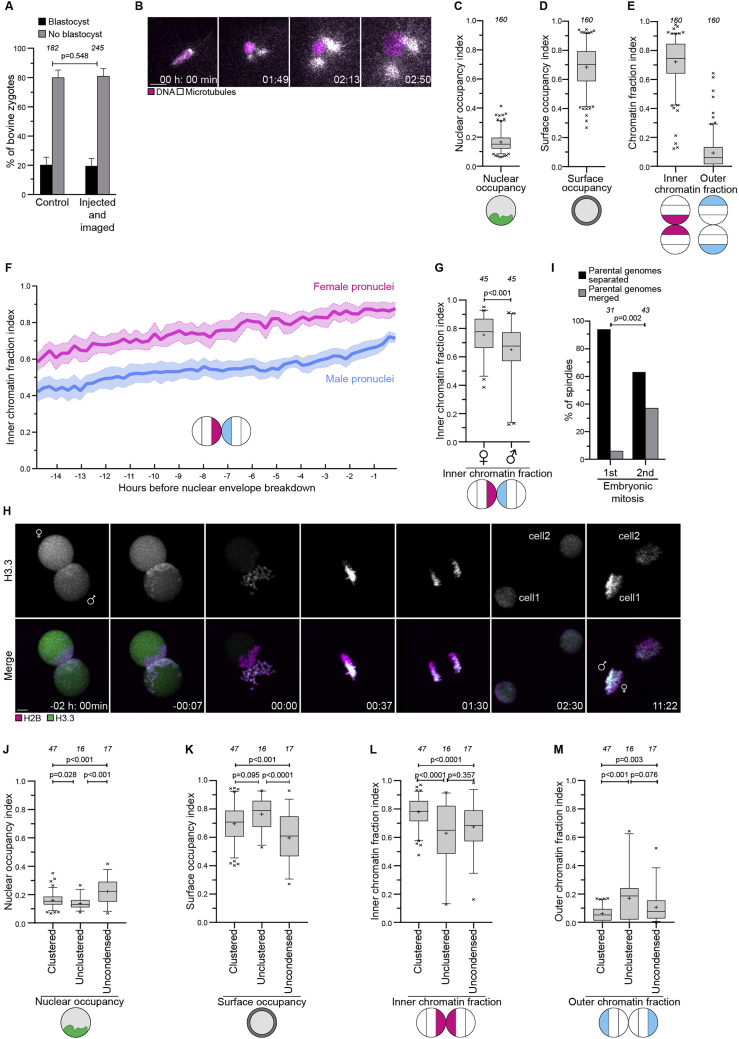
Figure S2.
The parental genomes cluster at the pronuclear interface in bovine zygotes, related to Figure 2
(A) Bovine zygotes non-injected and non-imaged or injected and imaged that developed into blastocysts or developed abnormally.
(B) Representative stills from time-lapse movies of male pronuclei imaged shortly after egg fertilization. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Time, h:min, 00:00 corresponds to start of image acquisition. First time point shows a single confocal section, while the other time points are Z-projections of 2 (01:49) or 4 (02:13 to 03:14) sections every 2.50 μm.
(C-E) Quantification of chromatin distribution within pronuclei before NEBD using the nuclear occupancy (I), surface occupancy (J), inner and outer chromatin fraction (K), indices.
(F) Inner chromatin fraction indices in female (magenta) and male (cyan) pronuclei. Solid lines represent means of five pronuclei belonging to five zygotes. Shaded areas represent standard error of the mean.
(G) Inner chromatin fraction indices in female (♂) and male (♀) pronuclei before NEBD.
(H) Representative stills from a time-lapse movie of a zygote undergoing cell division. Green, paternal DNA (H3.3-mClover3). Magenta, DNA (H2B-mScarlet). ♀ and ♂ indicate the female and the male pronucleus and the DNA of maternal and paternal origin in the second cell division. Time, h:min, 00:00 is NEBD. Z-projections, 10 sections every 2.50 μm. Scale bar, 10 μm.
(I) Embryos at the indicated stages were scored for spatial separation or merging of the parental genomes on the metaphase spindle.
(J-M) Quantification of chromatin distribution within pronuclei using the nuclear occupancy index (J), surface occupancy index (K), inner chromatin fraction index (L), and outer chromatin fraction index (M) in indicated groups in zygotes.
Data are from five (A, I), eleven (C, D, E, J, K, L, M), three (G), or seven (I) independent experiments. The number of analyzed zygotes (A), pronuclei (C, D, E, G, J, K, L, M), or spindles (I) is specified in italics. p values were calculated using Fisher’s exact test (A, I) and unpaired two-tailed Student’s t test (G, J, K, L, M). Scale bars, 10 μm.