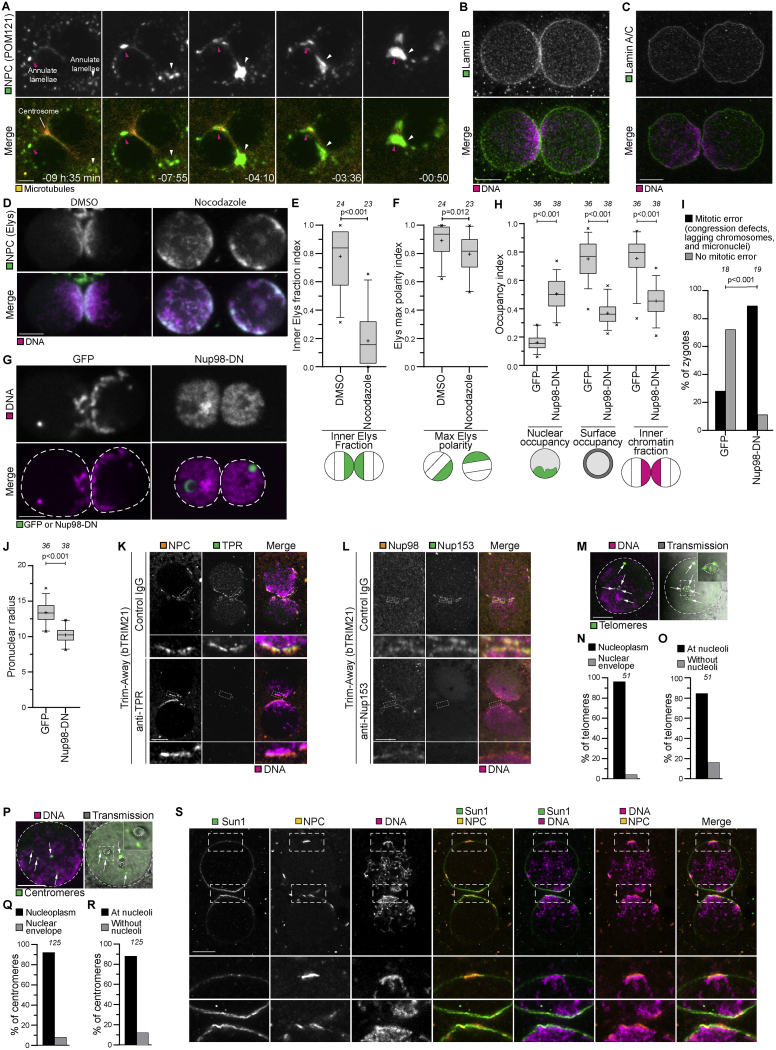
Figure S6.
Nuclear pore complexes cluster with chromatin at the pronuclear interface in bovine zygotes, related to Figure 5
(A) Representative stills from time-lapse movies of bovine zygotes expressing mClover3-MAP4-MTBD (microtubules, orange) and POM121-mScarlet (NPCs, green). Magenta and white arrowheads indicate two patches of annulate lamellae moving toward centrosomes and pronuclear interface. Time, h:min, 00:00 is NEBD. Single confocal microscopy sections.
(B-C) Representative immunofluorescence images of zygotes stained with Lamin B (B) or Lamin A/C (C) (Green) and DNA (DAPI, Magenta). Single sections Airyscan microscopy (B) and single sections confocal microscopy (C).
(D) Representative stills from time-lapse movies of zygotes expressing bElys-mClover3 (NPC, green) and H2B-mScarlet (DNA, magenta) treated with DMSO or nocodazole before pronuclear expansion. The time point at 4 hours before NEBD is shown. Z-projections of 2 (left) and 3 (right) sections every 2.50 μm.
(E-F) Inner Elys polarity and max Elys polarity indices at 4 hours before NEBD in zygotes treated with DMSO or nocodazole before pronuclear expansion. Analysis was not possible later because Elys redistributed to the chromatin before NEBD. Note that nocodazole was added after the formation of the midbody from the meiosis II spindle. Midbody microtubules are known to be very stable and, based on our experiments in parthenotes (Figures S7), might contribute to residual NPC polarization in nocodazole treated zygotes.
(G) Representative stills from time-lapse movies of zygotes before NEBD expressing mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN). Green, GFP or Nup98-DN. Magenta, DNA (H2B-mScarlet). Dashed lines mark pronuclei. Single section confocal microscopy.
(H) Nuclear occupancy, surface occupancy, and inner chromatin fraction indices in zygotes injected with mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN).
(I) Zygotes injected with mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN) having an abnormal mitosis.
(J) Pronuclear radius in zygotes injected with mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN).
(K-L) Representative immunofluorescence images of pronuclei in zygotes expressing bTrim21 (bovine Trim21) and treated with the indicated antibodies. Orange, NPC (NPC-mAb414 (K) or Nup98 (L)). Green, TPR (K) or Nup153 (L). Magenta, DNA (DAPI). Note that antibody injections were performed before fertilization to ensure protein depletion before pronuclear assembly.
(M) Representative immunofluorescence images of telomere distribution within a pronucleus. Gray, transmission. Magenta, DNA (DAPI). Green, telomeres (Trf1). Single sections confocal microscopy. Dashed lines indicate pronucleus and nucleoli. Arrows indicate telomeres. Outlined regions magnified in the top right corner.
(N) Telomeres in the nucleoplasm or at the nuclear envelope.
(O) Telomeres at nucleoli or away from nucleoli.
(P) Representative immunofluorescence images of centromere distribution within a pronucleus. Gray, transmission. Magenta, DNA (DAPI). Green, centromeres (ACA). Single sections confocal microscopy. Dashed lines indicate pronucleus and nucleoli. Arrows indicate centromeres. Outlined regions magnified in the top right corner.
(Q) Centromeres in the nucleoplasm or at the nuclear envelope.
(R) Centromeres at nucleoli or away from nucleoli.
(S) Representative immunofluorescence images of zygotes stained with Sun1 (Green), NPC (NPC-mAb414, Orange) and DNA (DAPI, Magenta). Outlined regions magnified in bottom two rows. Telomere bouquet formation during the early stages of meiosis relies on the LINC inner nuclear membrane protein SUN1. However, in bovine zygotes, SUN1 was distributed along both pronuclear envelopes, without specific enrichment on peripheral chromosomes (magnified region). This localization is in contrast to the clustered appearance of SUN1 in proximity to the centrosome in meiotic cells, but it is consistent with observations in C. elegans zygotes (Minn et al., 2009). Single sections Airyscan microscopy.
Data are from four (E, F) and five (H, I, J) independent experiments. Data in (N, O, Q, R) are from five embryos (two pronuclei each) generated in a single experiment. The number of analyzed pronuclei (E, F, H, J), zygotes (I), telomeres (N,O), and centromeres (Q, R) are specified in italics. p values were calculated using unpaired two-tailed Student’s t test (E, F, H, J) and Fisher’s exact test (I). Scale bars, 10 μm.