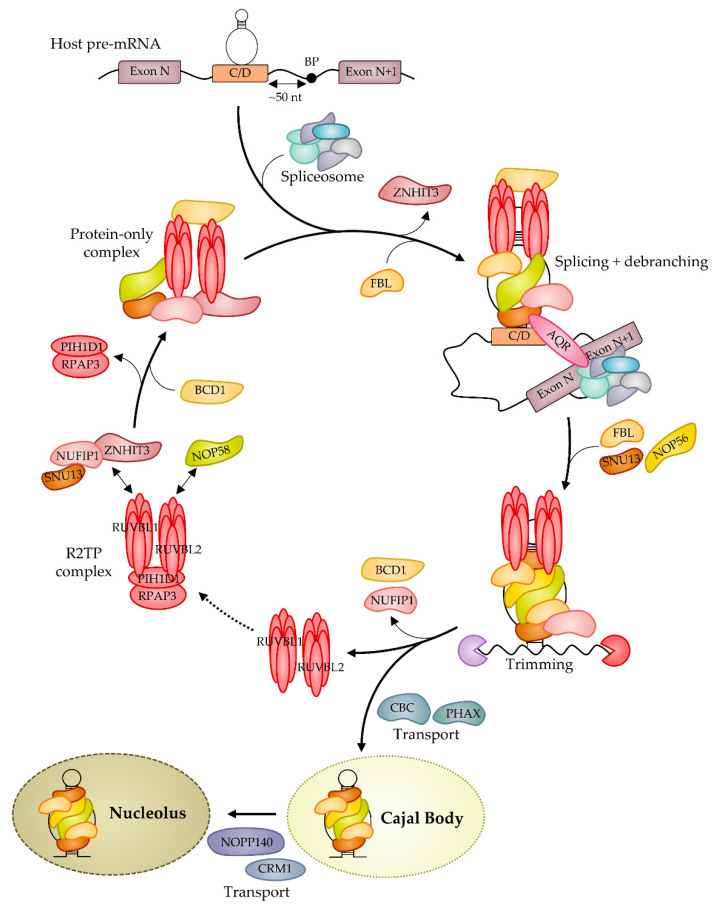
Figure 2.
Model of eukaryotic C/D snoRNP biogenesis. The loading of the core proteins SNU13, NOP58, NOP56, and FBL does not occur autonomously in eukaryotic cells but is mediated by several assembly factors. The R2TP complex contains the proteins RUVBL1, RUVBL2, RPAP3, and PIH1D1 and the latter interacts directly with NOP58 to stabilize it. The assembly platform NUFIP1 and its stabilizing factor ZNHIT3 interact with SNU13 to form a trimer. The essential contribution of the assembly factor BCD1 is less clear; it is integrated in a network of interactions involving PIH1D1, NUFIP1, RUVBL1&2, NOP58, and SNU13, which suggests that these proteins form a protein-only pre-snoRNP complex that scaffolds SNU13 and NOP58 core protein assembly. The proteins are then loaded on nascent snoRNA molecules in a co-transcriptional way upon the direct binding of SNU13 to the K-turn structures formed by the conserved C/D motifs. The efficient RNP assembly of intronic snoRNAs is favored by the association of the splicing and biogenesis machineries via the splicing factor AQR. The nucleation of the pre-snoRNP protects the snoRNA from extensive trimming by exonuclease activities. When released, the assembly factors might be recycled in a new biogenesis process (dotted line). Finally, the C/D snoRNPs are transported to CBs and nucleoli for functional purposes. This model is valid in both Saccharomyces cerevisiae and Human.