Abstract
Background
Urine from clinically healthy dogs is not sterile. Characterizing microbial diversity and abundance within this population of dogs is important to define normal reference ranges for healthy urine.
Objectives
To establish composition and relative representation of bacterial and fungal microbiomes in urine of clinically healthy dogs.
Animals
Fifty clinically healthy dogs.
Methods
Analytic study. Urine sampling via cystocentesis. Comprehensive evaluation of urine including standard urinalysis, culture and sensitivity, next‐generation sequencing (NGS), and bioinformatics to define bacterial and fungal microbiome.
Results
Culture did not yield positive results in any samples. Next‐generation sequencing of urine established low presence of bacteria, fungi, or both in all samples. Diversity and abundance of bacterial and fungal communities varied between urine samples from different dogs. Struvite crystals were associated with bacterial community structure (P = .07) and there was a positive correlation between struvite crystals and pH.
Conclusions and Clinical Importance
The microbiome in urine of clinically healthy dogs has diverse bacterial and fungal species These findings highlight limitations of conventional culture testing and the need for culture‐independent molecular diagnostics to detect microorganisms in urine.
Keywords: culture‐independent, microbiome, microbiota, mycobiome, urinalysis
Abbreviations
- ASV
amplicon‐sequence‐variant
- BCS
body condition score
- NGS
next‐generation sequencing
- PCA
principal component analysis
- PERMANOVA
permutational analysis of variance
- UTIs
urinary tract infections
1. INTRODUCTION
Improved understanding of the microbiome in health and disease has received increasing attention in human and veterinary medicine in the past decade and reliable reference data from clinically healthy individuals is thus needed. Differences exist in microbiome composition and abundance between individuals as well as anatomical sites. The urine microbiome was first established in clinically healthy humans around a decade ago. 1 Human urine is not sterile in both health and disease states. The bacterial urinary microbiome in clinically healthy dogs has greater taxonomic richness than rectal and genital microbiome. 2 Reference data is needed to enable evidence‐based differentiation between commensal and pathological microbes. Furthermore, recurrent urinary tract infections (UTIs) in companion dogs emphasize the importance of rapid and reliable microbiome monitoring over time. 2
Urine is an important health barometer with point‐of‐care screening focused on colorimetric dipstick, specific gravity, and microscopic testing. 3 Urine culture and sensitivity testing by a clinical laboratory typically involves a turn‐around time of 2 to 3 days, yet antibiotics are often prescribed empirically in cases with bacteriuria and clinical signs of UTI. Conventional urinalysis has limitations because many microorganisms remain undetected by culture methods, yielding “no growth” on culture despite signs of infection. 4 Lifetime prevalence of UTI in dogs is approximately 14% 5 ; however, clinical signs and culture results do not always match in individual cases. The ability to obtain more accurate test results quickly has therefore become a priority.
Next‐generation DNA sequencing (NGS) overcomes limitations of conventional culture‐based diagnostics by detecting and quantifying microbial DNA through untargeted and exhaustive sequencing and quantitative comparisons to reference databases. 6 NGS technology combined with bioinformatics is used increasingly to study both human and animal microbiomes in health and disease. Consequently, NGS‐based diagnostic tools are being adapted in veterinary medicine to optimize patient care and a microbial test is commercially available to detect and quantify aerobic and anaerobic bacteria and fungi. 7 The objective of the current study was to generate reference data on the bacterial and fungal canine urinary microbiome in “health.” This study presents comprehensive urine microbiome data in clinically healthy companion dogs.
2. MATERIALS AND METHODS
2.1. Sample collection and standard urinalysis
Clinically healthy dogs (n = 50) were prospectively identified by emailing a recruitment flyer to all faculty, staff, and students at Western University of Health Sciences (WesternU). The study was approved by the Institutional Animal Care and Use Committee at WesternU. Informed client consent was obtained and dogs were screened (ie, recording of medical history, physical exam, and urine collection) at the WesternU Pet Health Center during the Fall of 2019. One urine sample (6 mL) was collected via ultrasound‐guided cystocentesis with each dog in dorsal recumbency. Each urine sample was divided as follows: 2 mL into a BD Vacutainer Urinalysis tube (Becton, Dickinson and Company, Franklin Lakes, New Jersey) for standard urinalysis and 4 mL into a MiDOG Urine Collection tube (MiDOG LLC, Irvine, California) for urinalysis. MiDOG tubes contain 50 μL of urine conditioning buffer (Cat. No. D3061‐1‐140, Zymo Research Corp., Irvine, California) that preserves the microbial profile (microbiota) for prolonged periods at ambient temperature. 8 Consequently, all urine samples destined for microbiome analysis were batched and stored at 4°C until processing. Preserved samples were subsequently delivered to the MiDOG LLC testing facility (Irvine, California) and processed/sequenced as described in Section 2.2. Standard urinalysis was performed immediately after sample collection at an independent laboratory (ABID Diagnostics, Upland, California) and included visual exam, dipstick test, microscopic exam, and microbiology (aerobic culture and sensitivity). All urine samples were kept at 4°C processed within 12 hours after collection. Inclusion criteria included clinically healthy dogs without a history of antibiotic treatment for the preceding 6 months, current on vaccinations, spayed/neutered, body condition score (BCS) between 5 and 6, and negative urine aerobic culture results. Exclusion criteria included dogs treated with antibiotics, intact, not up‐to‐date on vaccinations, BCS below 5 or above 6, or positive urine culture.
2.2. DNA extraction and analysis of urine microbiota via NGS
The methods applied here were previously described. 7 Briefly, genomic DNA was purified using the ZymoBIOMICS‐96 DNA kit (Cat. No. D4304, Zymo Research Corp.) according to manufacturer's instructions in conjunction with a Hamilton Star liquid handling robot (Hamilton Company, Reno, Nevada). Zymo Research Corp. performed the sample library preparation and data analysis for both bacterial and fungal profiling (also the manufacturer of subsequent catalogue numbers, unless otherwise stated). Libraries were prepared using the Quick‐16S NGS Library Prep Kit (Cat. No. D6400) according to manufacturer's instructions with minor modifications. Primer sequences target the V1 to V3 region of the 16S rDNA and the ITS2 region for fungal analysis as previously described. 7 The exact sequences are proprietary to the MiDOG LLC service (https://www.midogtest.com). Libraries were sequenced using an Illumina HiSeq 1500 (Illumina, San Diego, California). Sequence reads were quality controlled using Dada2 (R package version 3.4). 9 Reads were trimmed and paired‐end reads were merged (perfect matching to primers, and no ambiguous nucleotides were allowed).
The microbiota profile of each sequenced urine sample was determined using the bioinformatics analysis pipeline offered by the MiDOG LLC testing service, which provides amplicon‐sequence‐variant (ASV) level (roughly species‐level) taxonomic identification. All phylotypes were computed as percent proportions based on the total number of sequences in each sample. The relative abundances of bacteria compared to fungi were determined assuming an equivalency of 1 16S rDNA copy to 1 fungal ITS (Internal Transcribed Spacers) copy. To control for any potential contamination of sequencing buffers, equipment, and other material, several negative controls were run for the extraction process as well as the library preparation. Specifically, for the “extraction negative control,” the storage buffer, DNA/RNA Shield (Cat. No. R1100‐50), was autoclaved at 120°C for 20 minutes and ZR BashingBead Lysis Tubes (Cat. No. S6012‐50) were baked at 120°C for 72 hours. One milliliter of autoclaved DNA/RNA Shield was added to baked ZR BashingBead Lysis Tubes (10 replicates). Control was bead beat on Vortex Genie 2 (SKU SI‐0236) equipped with horizontal tube adapter (SKU SI‐H524) at maximum speed for 40 minutes. The “negative extraction control” was lysed, extracted, library‐prepped, and sequenced in parallel with real experimental samples. The purpose of this negative extraction control was to assess the background of the workflow starting at the lysis step. For the “library preparation negative control,” ZymoBIOMICS DNA/RNAse free water (Cat. No. D4302‐5‐50) was autoclaved at 120°C for 20 minutes. Fifty microliters of autoclaved ZymoBIOMICS DNA/RNAse was added into empty wells (10 replicates) of the 96‐well plate containing the extracted DNA from experimental samples. This was the “no template control” for the library preparation process. This control was run in parallel with the DNA from experimental samples and extraction controls. The purpose of this “no template control” was to assess the background of the workflow starting from library preparation.
2.3. Bioinformatics and statistical analysis
Shapiro‐Wilks tests were used to test for normality of the data. Data tables generated by the MiDOG analytical pipeline were used for descriptive summaries and statistical hypothesis testing using the R software package. The number of reads for each ASV were summed for all of the negative controls and this total was subtracted from each of the samples (see also Table S1). In cases where this produced a negative number, the number of reads was recorded as 0. For initial descriptive statistics (shown in Figure 1), bacterial genera with fewer than 10 reads were removed from the analysis. For fungi, a minimum of 2 reads per taxon were required. For subsequent statistical analyses, because of the sparse nature of the data set, data were further screened to dogs with at least 3 bacterial genera detected (n = 39 dogs) and genera with a minimum of 10 reads and a mean relative abundance of at least 1% (n = 29 genera). For fungi, removing taxa with fewer than 2 reads resulted in a data set of 29 dogs by 7 genera. To identify significant clinical variables associated with variation in the number of bacterial genera (taxonomic richness) per dog, we used conditional inference trees implemented with the ctree command (part of the partykit library) in R (version 3.6.1). Conditional inference trees 10 are a nonparametric alternative to multiple regression approaches that use permutational binary partitioning to identify independent variables (clinical variables in our case, with Bonferroni corrections for multiple comparisons) that are significantly associated with the dependent variable (bacterial taxonomic richness). To determine the relationship between differences in bacterial community composition and struvite crystal levels for each dog, we combined principal component analysis (PCA) and permutational analysis of variance (PERMANOVA) both implemented in R with the prcomp and adonis functions (part of the stats and vegan packages), respectively.
FIGURE 1.
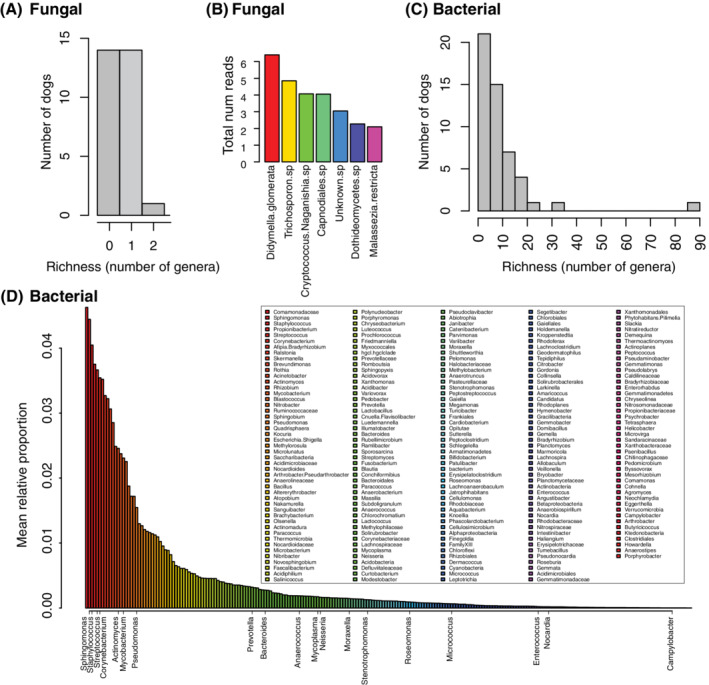
Summary of bacterial and fungal richness and community composition. A, Histogram of the number of fungal genera encountered per dog. B, Rank abundance of fungal taxa found across all dogs expressed as the total number of reads. C, Histogram of the number of bacterial genera encountered per dog. The minimum number of bacterial genera per dog was 1 (6 dogs), the median was 7, the mean 9.6, and the maximum 87. D, Rank abundance of bacterial genera across all dogs expressed as the mean relative abundance of each genus. As described in the Section 2, bacterial genera were screened for a minimum of 10 reads (n = 231) and fungal taxa for a minimum of 2 reads per taxon (n = 7). Legend for (D) shows all bacterial genera arranged in order of relative abundance from left to right across each column. Bacterial genera shown on x‐axis of (D) represent genera in the MiDOG database encountered in previous studies of the urine microbiome considered as known or potential pathogens
3. RESULTS
3.1. Evaluation of urine via conventional testing methods
All dogs enrolled in this study passed the inclusion criteria for clinically healthy dogs with no reported signs of UTI. The age of the dogs was 4.7 ± 3.0 years (mean ± SD) and 48% were female (24 of 50). Breeds represented in the study included, American pitbull terrier (4), German shepherd (3), Australian shepherd (2), American Staffordshire terrier (2), beagle (2), golden retriever (2), pug (2), Siberian husky (2), standard poodle (2), border collie (1), English bulldog (1), Great Dane (1), miniature American shepherd (1), vizsla (1), while the remaining 48% (24 of 50) were mixed‐breed. The body weight was 23.6 ± 11.1 kg (mean ± SD), including 66% (33 of 50) with a normal body score index. Most clinical urine variables were within an expected range of healthy individuals with some notable deviations (Table 1). Visual inspection of urine revealed haziness in 3 samples (6%). Specific gravity was within normal reference range (>1.030) in 94% (47 of 50) of dogs. Urine dipstick testing showed a pH between 6.0 and 7.5 in 64% (32 of 50) of the dogs with occasional evidence of proteinuria (12%, 6 of 50) and hematuria (12%, 6 of 50). Glucose, bilirubin, and ketones were within normal reference ranges. Microscopic analysis of urine samples showed sporadic erythrocytes (10%, 5 of 50) and leukocytes (4%, 2 of 50), with presence of epithelial cells or casts within normal limits. A total of 15 urine samples (30%, 15 of 50) contained struvite crystals to varying degrees (from “occasional” to “4plus”).
TABLE 1.
Data from standard urinalysis, including visual inspection, dipstick testing, light microscopy, and aerobic culture (n = 50 dogs)
| Color | Light yellow | Yellow | Dark yellow | ||||||
| 2 | 47 | 1 | |||||||
| Clarity | Clear | Cloudy | |||||||
| 48 | 2 | ||||||||
| Specific gravity | 1.037 (1.005‐1.056) | ||||||||
| pH | 6.6 (5.0‐9.0) | ||||||||
| Neg. | Trace | 1+ | 2+ | 3+ | 4+ | ||||
| Protein | 30 | 14 | 1 | 3 | 2 | 0 | |||
| Glucose | 49 | 1 | 0 | 0 | 0 | 0 | |||
| Ketones | 50 | 0 | 0 | 0 | 0 | 0 | |||
| Bilirubin | 49 | 0 | 0 | 1 | 0 | 0 | |||
| Hemoglobin | 37 | 7 | 2 | 1 | 1 | 2 | |||
| Leukocytes/HPF | None | Rare | 0‐3 | 3‐5 | 5‐10 | 25‐50 | 75‐100 | >100 | >>100 |
| 28 | 3 | 16 | 1 | 0 | 2 | 0 | 0 | 0 | |
| Erythrocytes | 29 | 4 | 6 | 6 | 1 | 1 | 1 | 1 | 1 |
| Epithelial cells | None | Rare | Few | 1+ | |||||
| 1 | 32 | 16 | 1 | ||||||
| Casts | None | Occasional | Granular 1+ | ||||||
| 47 | 2 | 1 | |||||||
| Crystals (struvite) | None | Occasional | 1+ | 2+ | 4+ | ||||
| 35 | 2 | 4 | 8 | 1 | |||||
| Culture | Negative | Positive | |||||||
| 50 | 0 | ||||||||
| Body condition score | 1‐4 | 5 | 6 | 7 | 8 | ||||
| 0 | 33 | 11 | 1 | 2 | |||||
| Age (y) | 4.7 ± 3.0 | ||||||||
| Weight (kg) | 23.6 ± 11.1 | ||||||||
Note: Mean, minimum, and maximum values are included for specific gravity and pH measurements; mean ± SD for age and body weight.
Abbreviation: HPF, high‐power field.
3.2. Microbial analysis of canine urine: Descriptive analyses
Aerobic microbial culturing was performed for each urine sample as described above. In all cases, the urine culturing result was “no growth.” DNA sequencing, however, amplified bacterial and or fungal taxa for each sample collected. After subtraction of reads from the negative controls as described in the Section 2, a total of 231 bacterial genera and 7 fungal species were identified across the entire sample set. Overall, the bacterial and fungal biomass and diversity were low in this population, with most dogs having from 1 to 10 bacterial genera (no dog had 0) and 1 or 0 fungal species (Figure 1A,C). The most abundant fungal taxa were Didymella glomerata, Trichosporon sp., and Cryptococcus naganishia (Figure 1B). The top 5 most dominant bacterial genera were Comamonadaceae (4.6% mean relative abundance), Sphingomonas (4.4%), Staphylococcus (4.0%), Propionibacterium (3.8%), and Streptococcus (3.7%) (Figure 1D). Interestingly, several genera commonly associated with known pathogens (or potentially opportunistic pathogens) were detected in the urine of clinically healthy individuals. These bacterial genera included Sphingomonas, 11 Staphylococcus, 12 , 13 Streptococcus, 14 Corynebacterium, 15 Actinomyces, 16 Mycobacterium, 17 Pseudomonas, 18 , 19 , 20 Stenotrophomonas, 21 Roseomonas, 22 Prevotella, 23 Nocardia, 24 , 25 Neisseria, 26 , 27 , 28 Mycoplasma, 29 , 30 , 31 Moraxella, 32 Micrococcus, 33 , 34 Enterococcus, 35 Campylobacter, 36 Bacteroides, 37 Anaerococcus, 38 and Actinomyces. 16 In addition, MiDOG LLC has detected Porphyromonas and Peptostreptococcus in a high number of urine samples from dogs with UTI (unpublished data), which is consistent with our findings (Figure 1D). These bacterial genera, considered as potential pathogens, were detected in anywhere from 1 to 17 dogs per genera, as shown in Table 2.
TABLE 2.
Bacterial genera identified in urine that might contain opportunistic pathogenic species
| Genus | Number of dogs |
|---|---|
| Streptococcus | 17 |
| Sphingomonas | 13 |
| Pseudomonas | 12 |
| Corynebacterium | 12 |
| Staphylococcus | 11 |
| Actinomyces | 7 |
| Mycobacterium | 6 |
| Porphyromonas | 4 |
| Moraxella | 4 |
| Bacteroides | 4 |
| Prevotella | 3 |
| Anaerococcus | 3 |
| Stenotrophomonas | 2 |
| Peptostreptococcus | 2 |
| Micrococcus | 2 |
| Fusobacterium | 2 |
| Roseomonas | 1 |
| Nocardia | 1 |
| Neisseria | 1 |
| Mycoplasma | 1 |
| Helicobacter | 1 |
| Finegoldia | 1 |
| Enterococcus | 1 |
| Campylobacter | 1 |
3.3. Linking the canine urine microbiome to clinical variables
Of the 20 clinical variables tested for significant associations with the number of bacterial genera (taxonomic richness) per dog using conditional inference trees as described in the methods, only struvite crystals were significant and this result was driven by a single dog in which 87 bacterial genera were detected with struvites characterized as “4plus.” This dog was bright, alert, responsive with normal appetite, no abnormalities in micturition pattern (no hematuria, pollakiuria, or dysuria) and considered clinically healthy. When this dog was removed from the data set as an outlier, no clinical variables tested had significant explanatory power for bacterial richness. To determine if taxonomic composition of the bacterial community varied according to clinical variables, we used PCA as described in the Section 2 to reduce the dimensionality of the community composition data and PERMANOVA to identify any significant correlations between the bacterial community composition and clinical variables. With the same data set as was used above to test for associations between taxonomic richness and clinical variables, only struvite crystal level was marginally significant according to analysis with PERMANOVA (Table 3). In the PCA, dogs with struvite levels classified as “2plus” were mostly differentiated from the other samples by the primary axis explaining 13% of the variation in community composition (Figure 2). Comparisons of struvite levels to other clinical variables showed that pH was a strong covariant with a pattern of increasing pH correlated with increasing struvite levels (Figure 3), but pH did not have a significant effect on community structure by itself or in combination with struvite levels as determined by PERMANOVA.
FIGURE 2.
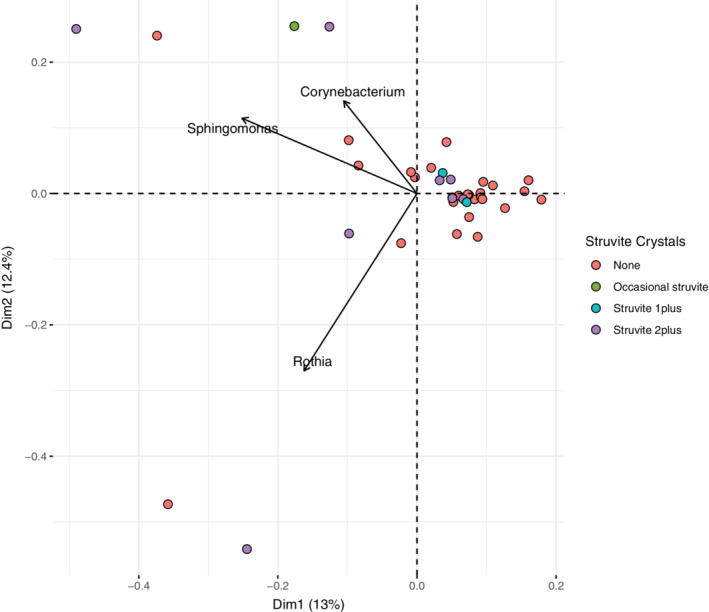
Analysis of beta‐diversity by PCA. Because of the sparse data, the data set was reduced to dogs with at least 3 bacterial genera detected (n = 39 dogs) and genera with a minimum of 10 reads and a mean relative abundance of at least 1% (n = 25 genera). Data points are colored according to struvite crystal levels based on the significance level shown in Table 3; arrows shows the magnitude and direction of the top 3 bacterial genera contributing to the clustering of the data. PCA, principal component analysis
FIGURE 3.
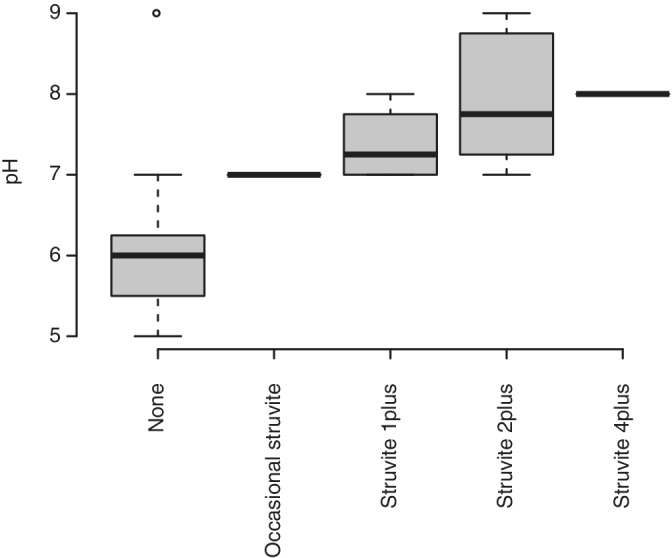
Relationship between pH values and Crystal levels for all 50 dogs
TABLE 3.
PERMANOVA results testing for significant relationships between clinical variables measured and bacterial community structure for data shown in Figure 2
| df | SUMOFSQS | R 2 | F | Pr(>F) | |
|---|---|---|---|---|---|
| Crystals | 3 | 1.5525 | 0.09646 | 1.2099 | 0.072 |
| Residual | 34 | 14.5417 | 0.90354 | ||
| Total | 37 | 16.0941 | 1.00000 |
Note: Of 20 variables tested (Age_yrs, Weight_kg, Urine_vol_ml, Specific_Gravity, pH, pH_level, Breed, Gender, Diet, BCS, Color, Appearance, Protein, Glucose, Blood, WBC, RBC, Epithelial_Cells, Casts, Crystals), only Crystals was marginally significant (P < .1). The Adonis function was used in R with the following command: adonis2(formula = subset.data ~ Crystals, data = meta.data.subset, permutations = 9999, method = “bray”).
Abbreviation: PERMANOVA, permutational analysis of variance.
Next, to identify bacterial genera with significantly different abundance between dogs grouped according to struvite crystal levels, we compared the relative abundance of bacterial genera according to the groupings—“none,” “occasional,” or “1plus” vs dogs with struvite crystals classified as “2plus” (Figure 4). The 3 bacterial genera shown in Figure 3 that were the top contributors to the PCA (Corynebacterium, Sphingomonas, and Rothia) had mean relative abundances that were 2 to 4 times greater in the high struvite vs low struvite dogs (Figure 4). Using DESeq2 as described in the Section 2, Ruminococcaceae was also significantly over‐represented in the high‐struvite group when the single dog in which 87 bacterial genera were detected with struvites characterized as “4plus” was included in the analysis. When this dog was removed from the analysis as an outlier Ruminococcaceae was no longer significantly different between the high and low struvite groups.
FIGURE 4.
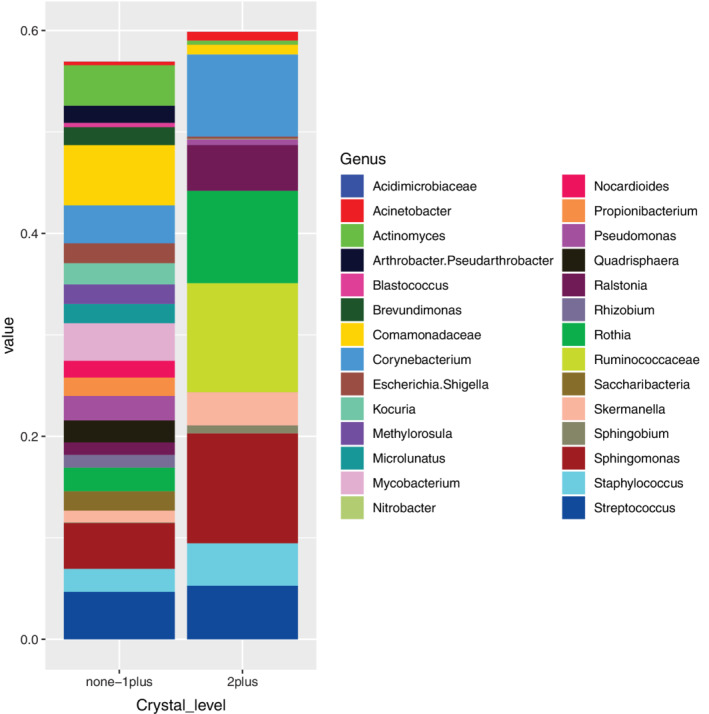
Comparison of mean relative abundance of bacterial genera according to struvite crystal level grouped into 2 groups according to PCA and PERMANOVA results. The same data set used for analysis of beta diversity in Figure 2 was used for this analysis. PERMANOVA, permutational analysis of variance
Although conditional inference trees, PCA, and PERMANOVA are nonparametric techniques and so do not require normally distributed data, we tested the continuous clinical data (age, weight, specific gravity, and pH) for normality—age and pH were positively skewed (Figure S1).
4. DISCUSSION
In this study, we confirm that urine from clinically healthy companion dogs is not sterile. Furthermore, we show that conventional urine culture might be inadequate for in‐depth clinical assessments of the urine microbiome. All dogs enrolled in this study had negative urine cultures; however, urinalysis using culture‐independent NGS revealed that all samples had bacteria present and that a diverse microbial community could be detected in many samples. It is noted that NGS cannot determine which microbes are viable in vivo. However, NGS can play an important role in nonculturable or unresolved chronic infections that are nonresponsive to antibiotic treatment prescribed according to culture and sensitivity results.
The development of NGS technology to study the associations of the microbiome with health status has drastically increased our understanding of the close interplay between the host and the microbiome. The majority of these efforts have been conducted in humans and to a much smaller extent on non‐human animal microbiomes. To date, relatively little is known about the canine urine microbiome in health and disease. Conventional teaching in veterinary medicine has stated that urine of healthy dogs is sterile; however, a recent study documented several bacterial species in urine collected via cystocentesis from clinically healthy dogs. 2 Data presented here demonstrate that urine from asymptomatic dogs can host a relatively rich and diverse microbial community including multiple taxa from at least 2 classes of microorganisms: fungi and bacteria. No previous study has shown the presence of fungi in the urine of clinically healthy dogs using an NGS approach. Future longitudinal studies of the microbiome from individual dogs could help deepen our understanding of the composition and function of the healthy microbiome and how it influences “normal” host physiology. Previous culture‐independent studies have also documented the nonsterile nature of urine samples from asymptomatic humans, 1 , 39 , 40 consistent with our results presented here (Figure 1). Importantly, these human data came from samples obtained through midstream urine collection, while the samples presented here were collected via cystocentesis. In veterinary medicine, it is universally accepted that the “free‐catch” method is less than ideal because the skin and external urogenital tissues are nonsterile areas that can serve as sources of contamination. 41 Therefore, this study could be considered to represent the microbiome of the bladder without “free‐catch”‐associated contamination of the microbiome profile. Future studies might consider including a “needle control” from the cystocentesis collection site of the skin as another negative control to further increase the study design rigor.
The 2019 diagnostic gold standard from the International Society for Companion Animal Infectious Disease 42 defines UTI based upon positive urine culture. In the present study, 100% of urine samples were culture negative (Table 1), all dogs were clinically healthy and urine samples were collected via cystocentesis, yet we detected a diverse urinary microbiome based on NGS. Urine metagenomic data from our sample of clinically healthy dogs identified a total of 231 bacterial genera, of which 20 have been characterized as potentially pathogenic (including Campylobacter, Pseudomonas, Actinomyces, and Staphylococcus) (Figure 1D and Table 2). This raises interesting questions regarding cell numbers or virulent phenotypes that might be required for these microorganisms to become opportunistic pathogens that are beyond the scope of the current study. Possible explanations for finding potentially pathogenic bacteria in clinically healthy dogs might include commonly observed false‐negative results for culture‐based diagnostics and the low threshold of detection for PCR‐based diagnostics. Our study design and bioinformatic workflows were careful to include multiple appropriate negative controls as described in the Section 2, for which any sequence reads were subtracted from all experimental samples.
Additionally, 7 fungal species were identified in the urine from across the sample set (Figure 1B), which is a new finding in veterinary medicine. The interrelationship between fungal and bacterial populations in canine urine has not been studied systematically to elucidate their roles in health and disease. As metagenomic data from dogs with UTIs becomes more widely available, we expect that the roles of fungi in the pathophysiology of UTIs might be better understood. The 7 different fungal taxa identified here are taxonomically diverse. Dothideomycetes (class) and D glomerata (species) are both Ascomycota, while Trichosporon (genus), Cryptococcus/Naganishia (genus), Malassezia restricta (species), and Capnodiales (order) are Basidiomycota. The fungal database is subject to several reclassifications over the past years, and D glomerata was previously known as both Phoma glomerata and Peyronellaea glomerata. 43 Didymella glomerata causes subcutaneous mycosis 44 and infection of the inner pinna in goats. 43 Several members of the order Capnodiales are opportunistic pathogens in dogs. 43
Importantly, these kinds of data might help reshape current therapeutic strategies by not only targeting bacteria identified using conventional culture and sensitivity testing, but also shed light on potentially synergistic roles of fungal species in urine. Our data show that numerous bacterial species seemingly coexist with a smaller number of fungal species in culture‐negative urine collected from clinically healthy dogs.
We tested the taxonomic richness of the urine microbiome from each dog against 20 clinically relevant variables (including breed, sex, weight, age, body score index, diet, and the 14 standard urinalysis variables obtained by visual inspection, dipstick testing, and microscopy as shown in Table 1) to identify any significant associations. Struvite crystals had the strongest association with bacterial richness and community structure (Figures 2 and 4). Clinically, the presence of this type of crystals in canine urine might be considered within normal limits; however, it has generally been regarded as a strong indicator of canine UTIs resulting from urease‐producing bacteria (eg, Staphylococcus spp., Proteus spp., Klebsiella spp., and Pseudomonas spp.). 45 , 46 Of 50 clinically healthy dogs, 1 individual had a high amount (“4plus”) of struvite crystals and the highest bacterial richness (87 genera), yet no clinical signs of UTI. The metagenomic analysis of the 10 most abundant bacteria present in the urine of this particular individual showed that at least five are considered urease‐producing bacteria (ie, Sporosarcina pasteurii, Viribacter sp., Mycobacterium sp., Rhodobiaceae sp., and Sporosarcina contaminans). None of these 5 bacterial species have been clinically associated with, or cultured from, healthy dogs or dogs with UTIs. This might explain the higher number of struvite crystals in urine from this particular dog. Sporosarcina pasteurii (formerly known as Bacillus pasteurii) has been investigated in nonmedical fields such as construction engineering due to its unique urease producing ability. 47 , 48 Sporosarcina pasteurii mutants created by UV irradiation have enhanced urease activity, calcite (calcium carbonate) precipitation, and survival at higher pH values. 47 Although the biology of this particular dog with struvite “4plus” is quite intriguing, we recognize this individual is an outlier. While it is important to recognize that no generalizable conclusions can be made regarding struvite crystals and urine microbiome from this single dog, data can serve as the foundation for a future pilot study on the association between struvite crystalluria and the urine microbiome. When we excluded this dog from the data set, none of the clinical variables tested had significant explanatory power for bacterial richness. Nevertheless, this dog was followed for more than a year after the urine sample collection and urinalysis and remained clinically healthy and free of any UTI signs. Subclinical bacteriuria occurs in dogs that have remained clinically healthy for 3 months after bacteriuria was noted, without any antimicrobial treatment. 49 Prevalence of asymptomatic bacteriuria in selected populations is low (2.1%) and that the presence of bacteria in urine with no associated clinical signs of UTI might be transient or persistent phenomenon. 50 Using pairwise comparisons of each clinical variables to struvite crystal classifications, we identified pH as a strong covariant with a clear pattern of increasing pH correlated with increasing struvite urine levels (Figure 3). These results are well aligned with current clinical knowledge that struvite crystals and uroliths are more efficiently formed in alkaline urine via urease‐induced production of ammonium, thus perpetuating the basic pH microenvironment. 51
Principal coordinate analysis was used to examine beta diversity (Figure 2), a metric of “differentiation diversity” between samples commonly used to understand how variation in community composition is correlated with environmental or clinical variables. 52 Beta diversity measures dissimilarity on a normalized scale from 0 to 1. As such, Bray‐Curtis dissimilarity equals 0 if samples have an identical microbial composition, and 1 if samples share no microbial species (ie, are completely different from one another). Total variance in a community composition matrix is used to estimate beta diversity indices, 53 where “community” refers to a group of potentially interacting microorganisms coexisting within space and time. As such, the reasoning for using PCA is to establish the degree of compositional dissimilarity in microbial community structure across all samples and determine if microbial diversity across the entire data set can be explained by any of the measured clinical variables. Figure 2 shows data in a 2‐dimensional space, representing the 2 primary axes explaining the variance of the dataset. Of all the clinical variables measured, only struvite crystal levels were significantly correlated with changes in the microbial community composition across samples. The first dimension (PC1) explained 13% of the total variance and was correlated with moderate (“2plus”) levels. The 3 bacterial genera that were the principal drivers of changes in the community structure (black arrows), were Corynebacterium, Rothia, and Sphingomonas, all of which were more abundant in the 2plus struvite dogs (Figure 4). With relatively few exceptions, data points tended to cluster around the origin (center point) of the graph, suggesting limited variability (ie, a high number of shared taxa) among samples.
When we compared the relative abundance of bacteria between the low‐ and high‐struvite groups and included the dog with the highest struvite load, we found 1 bacterial taxon, Ruminococcaceae, (part of the Clostridia) that was significantly overrepresented in urine from dogs with high‐struvite crystal load, that is, struvite 2plus (n = 8) and 4plus (n = 1). When the dog with the highest struvite load was excluded, this trend was still apparent (Figure 4), but not statistically significant. Anaerobic bacteria of the Ruminococcaceae family can be found in high numbers in the dog gut microbiota where they act as fiber fermenters. 54 To our knowledge, no peer‐reviewed reports have demonstrated the presence of Ruminococcaceae strains in normal (or infected) dog urine. However, other members of the bacterial order Rickettsiales, including Rickettsia, Ehrlichia, and Anaplasma, are vector‐borne pathogens. 55 , 56 , 57
A limitation of the study is that it solely focused on 1 geographical area (ie, dogs in Los Angeles County, California). Due to the lack of data regarding the healthy canine urine microbiome, this work was not hypothesis‐driven. Urine samples from dogs were collected via ultrasound‐guided cystocentesis after disinfecting the abdominal area with a sterile, alcohol‐soaked, gauze. However, we did not collect needle puncture samples from the skin surface to detect potential contamination with cutaneous microflora in the urine. The bacterial genera identified in the urine microbiome in our study differ significantly from the skin microbiome previously reported in clinically healthy dogs, 58 which makes contamination less likely. Based on the data presented here, we cannot definitively say if all of the taxa we identified came from viable microorganisms. It is well established that only a relatively small proportion of microorganisms are culturable with traditional methods, 4 , 59 , 60 but we still would have expected to culture several of the more common bacterial genera in our samples such as Escherichia coli, Proteus sp., or Klebsiella sp.
From a clinical perspective, it was surprising to find several hundred bacterial and more than half a dozen fungal species in urine from culture‐negative healthy adult dogs. However, our results are well aligned with recent evidence that urine samples from dogs without clinical signs of disease harbor at least 80 different bacterial strains. 2 Importantly, although the previous study used similar metagenomic techniques, it focused solely on bacterial taxa in normal dog urine without elucidating on fungal taxa. One explanation for negative culture results might be that the majority of the bacterial strains are in a “viable but nonculturable” state, 4 perhaps suppressed by the urine immune defense mechanisms in clinically healthy dogs or simply dormant due to unfavorable conditions. Future studies should use a variety of methods to distinguish viable from nonviable microbes.
CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
Approved by the WesternU IACUC (protocol # R19IACUC037) and client consent was obtained for all dogs enrolled in the study.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
Supporting information
Supplementary Figure 1 Normality tests for continuous clinical data showing frequency distributions and qq‐plots for each variable. Deviations from normality were tested with the Shapiro‐Wilks test; P‐values >0.05 indicate normally‐distributed data.
Supplementary Table 1 Additional information on the relative amplicon‐sequence‐variants (ASVs).
ACKNOWLEDGMENT
Funding for this study was provided by a grant to Tonatiuh Melgarejo and Brian B. Oakley from the WesternU College of Veterinary Medicine. MiDOG LLC sponsored microbiome analyses and client incentives for enrollment. The authors thank nursing and technical staff at the WesternU Pet Health Center and students who enrolled their dogs in the study. We thank Barrak Pressler for reviewing the manuscript.
Melgarejo T, Oakley BB, Krumbeck JA, Tang S, Krantz A, Linde A. Assessment of bacterial and fungal populations in urine from clinically healthy dogs using next‐generation sequencing. J Vet Intern Med. 2021;35:1416–1426. 10.1111/jvim.16104
Funding information WesternU College of Veterinary Medicine
REFERENCES
- 1. Kogan MI, Naboka YL, Ibishev KS, Gudima IA, Naber KG. Human urine is not sterile ‐ shift of paradigm. Urol Int. 2015;94:445‐452. [DOI] [PubMed] [Google Scholar]
- 2. Burton EN, Cohn LA, Reinero CN, Rindt H, Moore SG, Ericsson AC. Characterization of the urinary microbiome in healthy dogs. PLoS One. 2017;12:e0177783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Byron JK. Urinary tract infection. Vet Clin North Am Small Anim Pract. 2019;49:211‐221. [DOI] [PubMed] [Google Scholar]
- 4. Barcina I, Arana I. The viable but nonculturable phenotype: a crossroads in the life‐cycle of non‐differentiating bacteria. Rev Environ Sci Biotechnol. 2009;8:245‐255. [Google Scholar]
- 5. Ling GV. Therapeutic strategies involving antimicrobial treatment of the canine urinary tract. J Am Vet Med Assoc. 1984;185:1162‐1164. [PubMed] [Google Scholar]
- 6. Krumbeck J, Linde A, Melgarejo T. Clinical applications of next‐generation sequencing and microbiome analysis: the solution to no‐growth results. Pulse. 2019; November, 19‐20. [Google Scholar]
- 7. Tang S, Prem A, Tjokrosurjo J, et al. The canine skin and ear microbiome: a comprehensive survey of pathogens implicated in canine skin and ear infections using a novel next‐generation‐sequencing‐based assay. Vet Microbiol. 2020;247:108764. [DOI] [PubMed] [Google Scholar]
- 8. Keller D, Rothen J, Dangy JP, et al. Performance of a real‐time PCR approach for diagnosing Schistosoma haematobium infections of different intensity in urine samples from Zanzibar. Infect Dis Poverty. 2020;9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat. 2006;15:651‐674. [Google Scholar]
- 11. Oh YI, Kim HJ, Kim YM, et al. Antimicrobial resistance of bacterial isolates from positive urine culture in four hundred five dogs between 2013‐2014. Int J Appl Res Vet Med. 2017;15:99‐107. [Google Scholar]
- 12. Penna B, Varges R, Martins R, et al. In vitro antimicrobial resistance of staphylococci isolated from canine urinary tract infection. Can Vet J. 2010;51:738‐742. [PMC free article] [PubMed] [Google Scholar]
- 13. Rubin JE, Gaunt MC. Urinary tract infection caused by methicillin‐resistant staphylococcus pseudintermedius in a dog. Can Vet J. 2011;52:162‐164. [PMC free article] [PubMed] [Google Scholar]
- 14. Lamm CG, Ferguson AC, Lehenbauer TW, Love BC. Streptococcal infection in dogs: a retrospective study of 393 cases. Vet Pathol. 2010;47:387‐395. [DOI] [PubMed] [Google Scholar]
- 15. Maurey C, Boulouis HJ, Canonne‐Guibert M, Benchekroun G. Clinical description of Corynebacterium urealyticum urinary tract infections in 11 dogs and 10 cats. J Small Anim Pract. 2019;60:239‐246. [DOI] [PubMed] [Google Scholar]
- 16. Jacob ME, Crowell MD, Fauls MB, Griffith EH, Ferris KK. Diagnostic accuracy of a rapid immunoassay for point of‐care detection of urinary tract infection in dogs. Am J Vet Res. 2016;77:162‐166. [DOI] [PubMed] [Google Scholar]
- 17. Wiebe VJ. Chapt. 95: mycobacterium species. Section C: therapy of established infections. Valerie J. Wiebe. Drug Therapy for Infectious Diseases of the Dog and Cat. Hoboken, New Jersey: Wiley; 2015:90 [Google Scholar]
- 18. Bowles MH, Welsh RD, Hoffman J, Turnwald GH. Evaluation of a method using Proteus mirabilis and Pseudomonas aeruginosa to experimentally induce dual infection of the urinary bladder in dogs. Am J Vet Res. 2000;61:1484‐1486. [DOI] [PubMed] [Google Scholar]
- 19. Thompson MF, Litster AL, Platell JL, Trott DJ. Canine bacterial urinary tract infections: new developments in old pathogens. Vet J. 2011;190:22‐27. [DOI] [PubMed] [Google Scholar]
- 20. Hariharan H, Sylvester EB, Matthew V. Antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa from dogs in Grenada. West Indian Vet J. 2009;9:1‐3. [Google Scholar]
- 21. Kralova‐Kovarikova S, Husnik R, Honzak D, Kohout P, Fictum P. Stenotrophomonas maltophilia urinary tract infections in three dogs: a case report. Vet Med. 2012;57:380‐383. [Google Scholar]
- 22. Singh BR. Urinary Tract Infections (UTIs): The Most Common Causes and Effective Antimicrobials. Izatnagar, India: Division of Epidemiology, Indian Veterinary Research Institute; 2019. [Google Scholar]
- 23. Ling GV, Norris CR, Franti CE, et al. Interrelations of organism prevalence, specimen collection method, and host age, sex, and breed among 8,354 canine urinary tract infections (1969‐1995). J Vet Intern Med. 2001;15:341‐347. [PubMed] [Google Scholar]
- 24. Poisnel E, Roseau JB, Landais C, Rodriguez‐Nava V, Bussy E, Gaillard T. Nocardia veterana: disseminated infection with urinary tract infection. Braz J Infect Dis. 2015;19:216‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yaemsiri S, Sykes JE. Successful treatment of disseminated Nocardiosis caused by Nocardia veterana in a dog. J Vet Intern Med. 2018;32:418‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alamri Y, Keene A, Pithie A. Acute cystitis caused by commensal Neisseria oralis: a case report and review of the literature. Infect Disord Drug Targets. 2017;17:64‐66. [DOI] [PubMed] [Google Scholar]
- 27. Lowy FD, Pollack S, Fadl‐Allah N, Steigbigel NH. Susceptibilities of bacterial and fungal urinary tract isolates to desferrioxamine. Antimicrob Agents Chemother. 1984;25:375‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Péc J, Moravcík P, Kliment J, et al. Isolation of Neisseria gonorrhoeae from urine obtained by suprapubic puncture of bladders of men with gonococcal urethritis. Genitourin Med. 1988;64:156‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hemmatzadeh F, Niap F, Bennett BA, Trott DJ, Peaston AE. A novel quantitative polymerase chain reaction to monitor urinary tract mycoplasma infection in a dog. Lett Appl Microbiol. 2019;68:409‐414. [DOI] [PubMed] [Google Scholar]
- 30. Jang SS, Ling GV, Yamamoto R, Wolf AM. Mycoplasma as a cause of canine urinary tract infection. J Am Vet Med Assoc. 1984;185:45‐47. [PubMed] [Google Scholar]
- 31. Sykes JE, . Mycoplasma infections. In: Sykes JE, ed. Canine and Feline Infectious Diseases‐E‐Book. 1st. St. Louis, Missouri: Elsevier Health Sciences; 2013:382‐389. [Google Scholar]
- 32. Wiebe VJ. Chapt. 26: urinary tract infections. Drug Therapy for Infectious Diseases of the Dog and Cat. 1. Ames, Iowa: Wiley‐Blackwell; 2015:22. [Google Scholar]
- 33. Elbashier AM, Deshpande H. Recurrent urinary tract infection with haematuria caused by Moraxella (Branhamella) catarrhalis. J Infect. 1993;27:216‐217. [DOI] [PubMed] [Google Scholar]
- 34. Cetin C, Senturk S, Kocabiyik AL, et al. Bacteriological examination of urine samples from dogs with symptoms of urinary tract infection. Turk J Vet Anim Sci. 2003;27:1225‐1229. [Google Scholar]
- 35. Marques C, Belas A, Franco A, Aboim C, Gama LT, Pomba C. Increase in antimicrobial resistance and emergence of major international high‐risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J Antimicrob Chemother. 2018;73:377‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krunoslav B. Campylobacter Species in Dogs and Cats and Significance to Public Health in New Zealand: A Thesis in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy in Veterinary Science at Massey University, Palmerston North, New Zealand; 2016.
- 37. Ball KR, Rubin JE, Chirino‐Trejo M, Dowling PM. Antimicrobial resistance and prevalence of canine uropathogens at the Western College of Veterinary Medicine Veterinary Teaching Hospital, 2002‐2007. Can Vet J. 2008;49:985‐990. [PMC free article] [PubMed] [Google Scholar]
- 38. Säemann M, Hörl WH. Urinary tract infection in renal transplant recipients. Eur J Clin Invest. 2008;38(suppl 2):58‐65. [DOI] [PubMed] [Google Scholar]
- 39. Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Price TK, Dune T, Hilt EE, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54:1216‐1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barsanti JA. Genitourinary infections. In: Greene CE, ed. Infectious Diseases of the Dog and Cat. St. Louis, MO: Saunders Elsevier; 2006:935‐961. [Google Scholar]
- 42. Weese JS, Blondeau J, Boothe D, et al. International Society for Companion Animal Infectious Diseases (ISCAID) guidelines for the diagnosis and management of bacterial urinary tract infections in dogs and cats. Vet J. 2019;247:8‐25. [DOI] [PubMed] [Google Scholar]
- 43. Hurst CJ. Dirt and disease: the ecology of soil fungi and plant fungi that are infectious for vertebrates. In: Hurst Christian J, ed. Advances in Environmental Microbiology: Understanding Terrestrial Microbial Communities. 1. Cham, Switzerland: Springer International Publishing; 2019:289‐405. [Google Scholar]
- 44. Stchigel AM, Sutton DA. Coelomycete fungi in the clinical lab. Curr Fungal Infect Rep. 2013;7:171‐191. [Google Scholar]
- 45. Armbruster CE, Smith SN, Johnson AO, et al. The pathogenic potential of Proteus mirabilis is enhanced by other uropathogens during polymicrobial urinary tract infection. Infect Immun. 2017;85(2):e00808‐e00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dheyab AS, Aljumaili OI, Hussein NM. Study of virulence factors in urease‐positive bacteria isolated from urinary tract infections clinical specimens. J Pure Appl Microbiol. 2018;12:1465‐1472. [Google Scholar]
- 47. Achal V, Mukherjee A, Basu PC, Reddy MS. Strain improvement of Sporosarcina pasteurii for enhanced urease and calcite production. J Ind Microbiol Biotechnol. 2009;36:981‐988. [DOI] [PubMed] [Google Scholar]
- 48. Omoregie AI, Khoshdelnezamiha G, Senian N, Ong DEL, Nissom PM. Experimental optimisation of various cultural conditions on urease activity for isolated Sporosarcina pasteurii strains and evaluation of their biocement potentials. Ecol Eng. 2017;109:65‐75. [Google Scholar]
- 49. Wan SY, Hartmann FA, Jooss MK, Viviano KR. Prevalence and clinical outcome of subclinical bacteriuria in female dogs. J Am Vet Med Assoc. 2014;245:106‐112. [DOI] [PubMed] [Google Scholar]
- 50. McGhie JA, Stayt J, Hosgood GL. Prevalence of bacteriuria in dogs without clinical signs of urinary tract infection presenting for elective surgical procedures. Aust Vet J. 2014;92:33‐37. [DOI] [PubMed] [Google Scholar]
- 51. Mawby DI, Callens A, Bartges JW. Top ten urology consult questions (chapter 203) in section IX: urinary diseases. In: Bonagura JD, Twedt DC, eds. Kirk's Current Veterinary Therapy XV ‐ Small Animal Practice. 1st ed. Philadelphia, PA: Saunders; 2014:223‐227. [Google Scholar]
- 52. Nemergut DR, Schmidt SK, Fukami T, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77:342‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ricotta C. Of beta diversity, variance, evenness, and dissimilarity. Ecol Evol. 2017;7:4835‐4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmitz S, Suchodolski J. Understanding the canine intestinal microbiota and its modification by pro‐, pre‐ and synbiotics ‐ what is the evidence. Vet Med Sci. 2016;2:71‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blanton LS. Rickettsiales: treatment and management of human disease. In: Thomas S, ed. Rickettsiales. Cham, Switzerland: Springer International Publishing; 2016:109‐124. [Google Scholar]
- 56. Campos‐Calderón L, Ábrego‐Sánchez L, Solórzano‐Morales A, et al. Molecular detection and identification of Rickettsiales pathogens in dog ticks from Costa Rica. Ticks Tick Borne Dis. 2016;7:1198‐1202. [DOI] [PubMed] [Google Scholar]
- 57. Schmutzhard E, Helbok R. Chapter 96: Rickettsiae, protozoa, and opisthokonta/metazoa. In: Biller J, Ferro JM, eds. Handbook of Clinical Neurology. (3rd series) Neurologic Aspects of Systemic Disease Part III. Vol 121. Amsterdam, The Netherlands: Elsevier B.V.; 2014:1403‐1443. [Google Scholar]
- 58. Rodrigues Hoffmann A, Patterson AP, Diesel A, et al. The skin microbiome in healthy and allergic dogs. PLoS One. 2014;9:e83197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369‐394. [DOI] [PubMed] [Google Scholar]
- 60. Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321‐346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Normality tests for continuous clinical data showing frequency distributions and qq‐plots for each variable. Deviations from normality were tested with the Shapiro‐Wilks test; P‐values >0.05 indicate normally‐distributed data.
Supplementary Table 1 Additional information on the relative amplicon‐sequence‐variants (ASVs).


