Thermodynamic parameters of 2′-OCF3 modified RNA obtained by UV melting profile analysis (including unmodified and 2′-SCF3 modified reference RNAs)a.
| Sequence (5′ → 3′) | T m [°C] | ΔG°298 [kcal mol−1] | ΔH° [kcal mol−1] | ΔS° [cal mol−1 K−1] |
|---|---|---|---|---|
| GGUCGACC | 59.1 | −14.3 | −72.0 | −193 |
| GGUCGA̲CC | 52.7 | −12.6 | −69.1 | −189 |
| GAAGG-GCAA-CCUUCG | 73.3 | −6.0 | −46.0 | −134 |
| GAA̲GG-GCAA-CCUUCG | 70.3 | −5.8 | −46.7 | −137 |
GA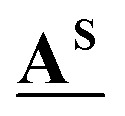 GG-GCAA-CCUUCG30 GG-GCAA-CCUUCG30
|
57.4 | −5.5 | −57.9 | −176 |
| GAAGG-GCAA-CC̲UUCG | 66.8 | −4.9 | −41.7 | −124 |
GAAGG-GCAA-C UUCG29 UUCG29
|
53.3 | −4.4 | −50.4 | −154 |
| GAAGG-GCA̲A-CCUUCG | 74.0 | −8.3 | −60.9 | −177 |
A̲ 2′-OCF3 adenosine, C̲ 2′-OCF3 cytidine, 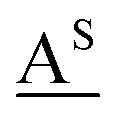 2′-SCF3 adenosine,30
2′-SCF3 adenosine,30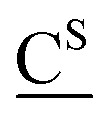 2′-SCF3 cytidine.29 Buffer: 10 mM Na2HPO4, 150 mM NaCl, pH 7.0. ΔH and ΔS values were obtained by van't Hoff analysis according to ref. 37 and 38. Errors for ΔH and ΔS, arising from non-infinite cooperativity of two-state transitions and from the assumption of a temperature-independent enthalpy, are typically 10–15%. Additional error is introduced when free energies are extrapolated far from melting transitions; errors for ΔG are typically 3–5%.
2′-SCF3 cytidine.29 Buffer: 10 mM Na2HPO4, 150 mM NaCl, pH 7.0. ΔH and ΔS values were obtained by van't Hoff analysis according to ref. 37 and 38. Errors for ΔH and ΔS, arising from non-infinite cooperativity of two-state transitions and from the assumption of a temperature-independent enthalpy, are typically 10–15%. Additional error is introduced when free energies are extrapolated far from melting transitions; errors for ΔG are typically 3–5%.
