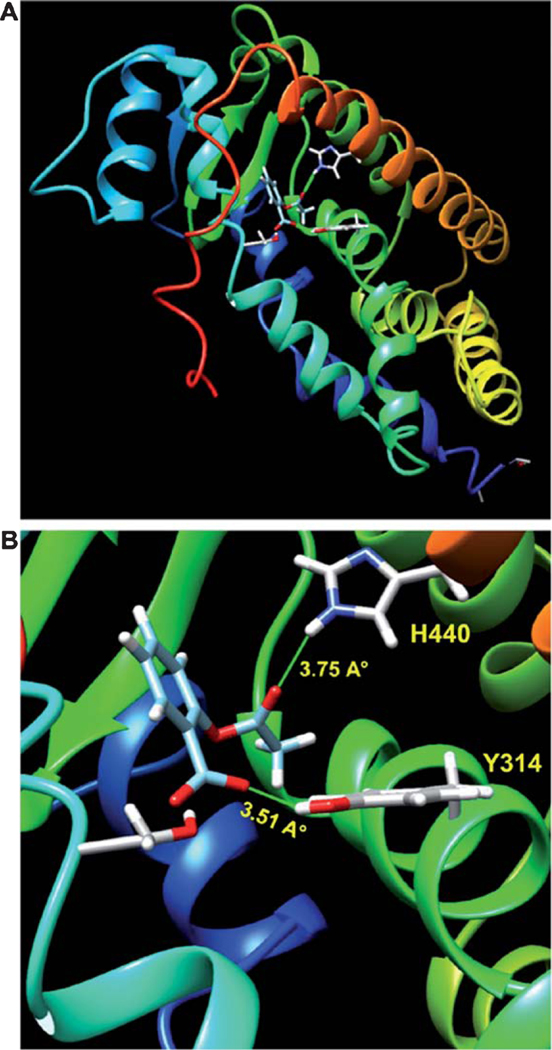
Fig. 1.
In Silico docking analysis of aspirin in PPARα ligand-binding domain (LBD). A, B) A best-fit superposition of the LBD of PPARα (mouse) and aspirin was obtained with the help of SwissDock docking software and finally displayed in Chimera visualization interface. Ribbon representation of the complex has revealed that a catalytic triad of H440, S280, and Y314 is required for the complex formation of aspirin with PPARα. Interestingly, hydrogen atom of OH group located at the sidechain of Tyr314 residue forms a moderate H-bond with O3 of Aspirin, whereas HE2 of H440 makes a weaker H-bond with O1 of Aspirin.