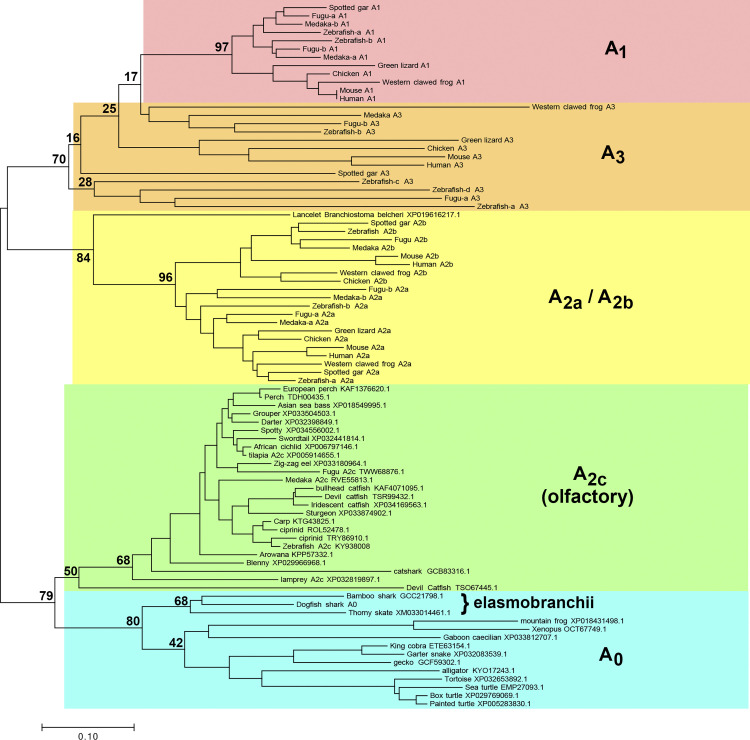
Figure 1.
Phylogenetic relationships of adenosine receptor (ADOR) taxa. Evolutionary history is inferred with the neighbor-joining method (14). The optimal tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the tree. Distances were calculated with the Poisson correction method (36) and are in the units of the number of amino acids substitutions per site. A1, A2a, A2b, A3, and olfactory (A2c) ADORs shown in Wakisaka et al. (13) are shown, using the same names as in that paper. The first characterized A2c (zebrafish, KY938008) by Wakisaka et al. (13) is also shown. Additional putative ADORs found in this work are shown by species and accession. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test using 500 replicates (40) are shown at selected branchpoints. Analyses were conducted with MEGA7 (18), using only full-length sequences for all receptors.