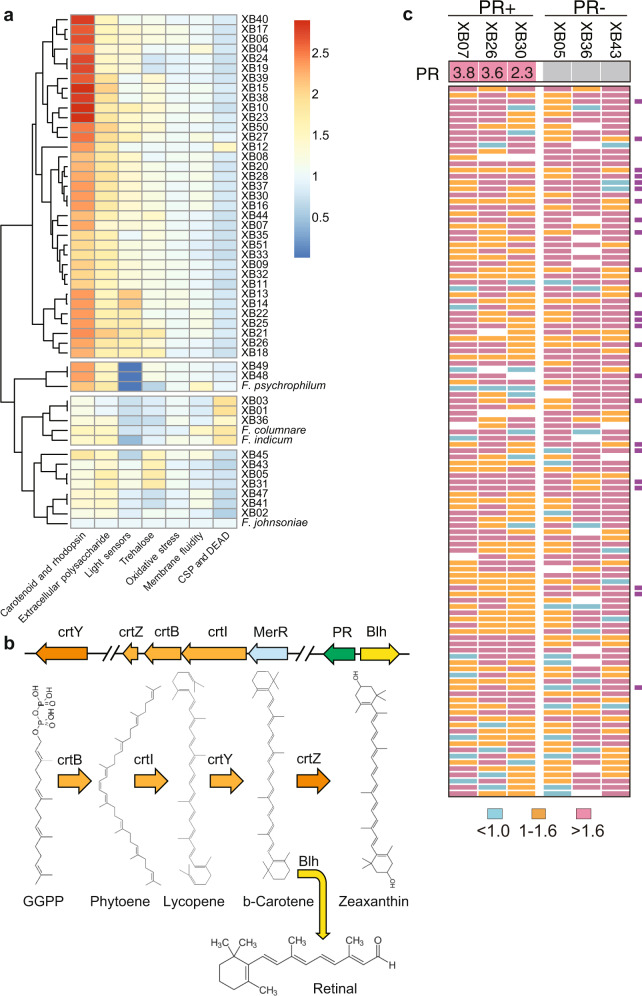
Fig. 5. Overrepresented gene categories in the glacial Flavobacterium genomes and the light-responding genes.
a Hierarchically clustered dendrogram with corresponding heatmap showing the relative abundances of the overrepresented gene categories among the 47 glacier strains. Color intensity represents the abundance fold difference of the corresponding genes in the genomes of the glacial Flavobacterium strains over that of F. johnsoniae, the type species of the genus. Blue and red coloring represent the minima and maxima abundance fold differences, respectively. Gene categories that may be related to cold adaptation are shown beneath each column. b A schematic shows gene organization of the crtIBZY operon that encodes enzymes for synthesizing zeaxanthin, proteorhodopsin (PR), and β-carotene dioxygenase (Blh) in the upper panel. The lower panel shows the predicted pathways of zeaxanthin and retinal synthesis in the glacial strains. c Heatmap showing the consensus upregulated genes in each of the three glacial prd-containing (PR+) and prd-lacking (PR−) Flavobacterium strains in light vs. dark cultures. Colored blocks show the fold changes of transcripts, as indicated beneath the heatmap. Numbers at the top of the heatmap indicate the fold changes of the prd gene (PR) expression in light vs. in dark cultures, as measured with quantitative RT-PCR. The purple blocks at the right specify the genes that primarily responded to light illumination, that are shadowed in Supplementary Dataset 2.