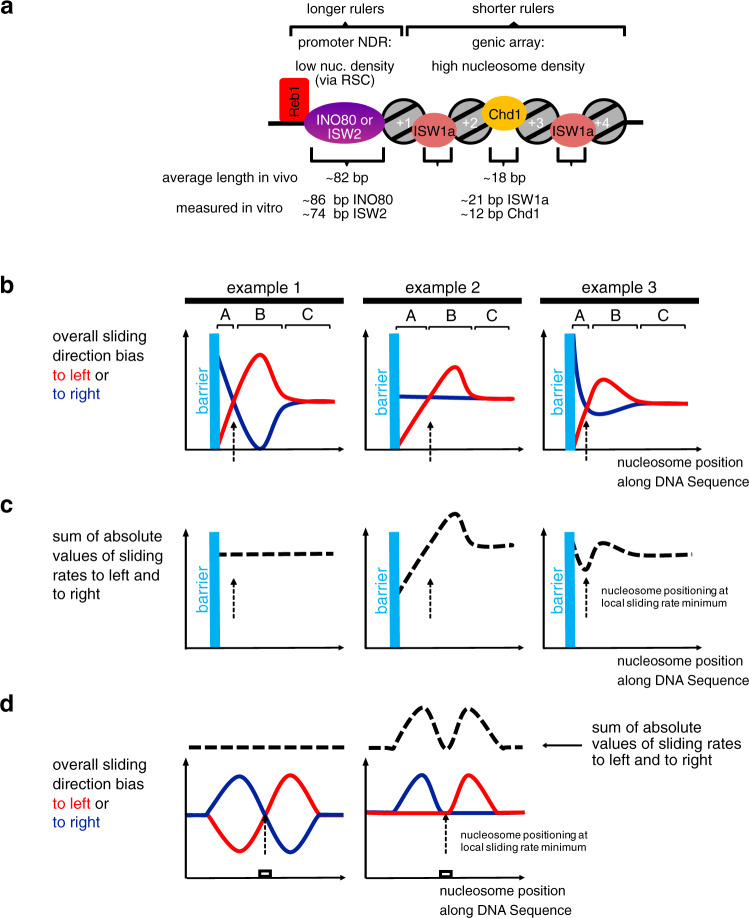
Fig. 7. Models for in vivo role of remodeler rulers and for ruler mechanism.
a Schematic showing how different remodelers contribute to the stereotypical nucleosome organization at 5′ ends of genes in vivo. The promoter is a nucleosome-depleted region (NDR) with low nucleosome density, e.g., due to nucleosome eviction by RSC. The gene body is organized in densely packed and regular nucleosomal arrays. The in vivo observed average distance between the flank of the +1 nucleosome and the center of a Reb1-binding site (distance to barrier) and the average linker length (spacing) mainly reflect the contribution to varying degrees of INO80’s or ISW2’s long ruler or of ISW1a’s or Chd1’s short ruler elements, respectively, as determined in vitro for low or high nucleosome density, respectively, in our study. This explains the main trends, while variations in remodeler type, nucleosome density as well as transcription, replication, and epigenetic marks may further locally modulate nucleosome organization. b Three hypothetical examples for how a remodeler ruler regulates the overall bias of sliding a nucleosome to left (red curves) or to right (blue curves) resulting in nucleosome positioning (stippled vertical arrows) in the vicinity of a barrier. c As b, but plotting sum of absolute values of sliding rates to the left and to the right (stippled black curves). d Two hypothetical examples for how a remodeler ruler leads to nucleosome positioning over a DNA sequence element (white box). Symbolics as in b and c. For details see text.