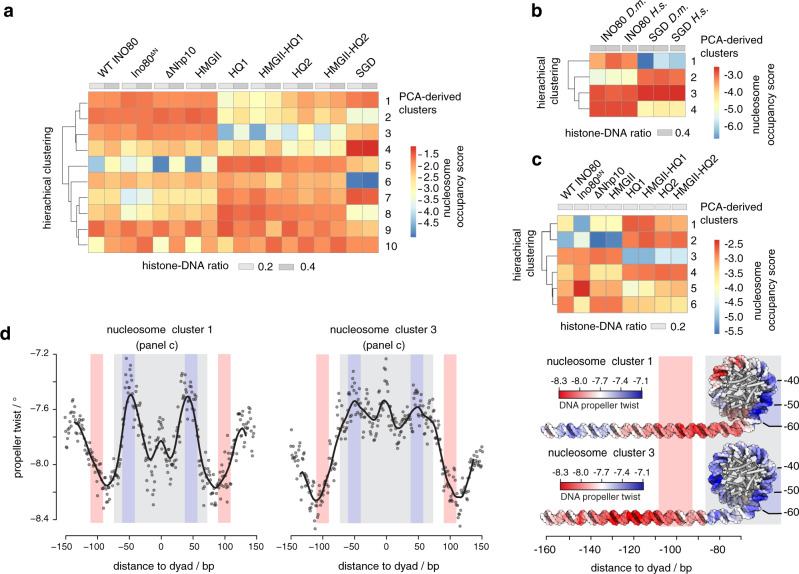
Fig. 5. Structure-based mutations probe the DNA shape/mechanics readout by INO80.
a Heat map representation of PCA/clustering analysis as in Fig. 3b, but not row scaled and for SGD chromatin with embryonic D. melanogaster histones at the indicated histone-to-DNA mass ratios either without (SGD) or after remodeling by the indicated recombinant S. cerevisiae WT and mutant INO80 complexes (mutants defined in Fig. 2c; for paired-end sequencing datasets see Supplementary Data 2 and GEO deposition at GSE145093.) b As a but for SGD chromatin with embryonic D. melanogaster (D.m.) versus recombinant H. sapiens (H.s.) histones and only remodeling by recombinant S. cerevisiae WT INO80 complex (INO80). c As a but only for the indicated subset of samples. d Left: as Fig. 4a but only for nucleosomes from the indicated clusters of c. Right: as in Fig. 4b, propeller twist DNA shape data were mapped onto model of linker and nucleosomal DNA by using red–white–blue color gradient. See Supplementary Fig. 3 for all clusters.