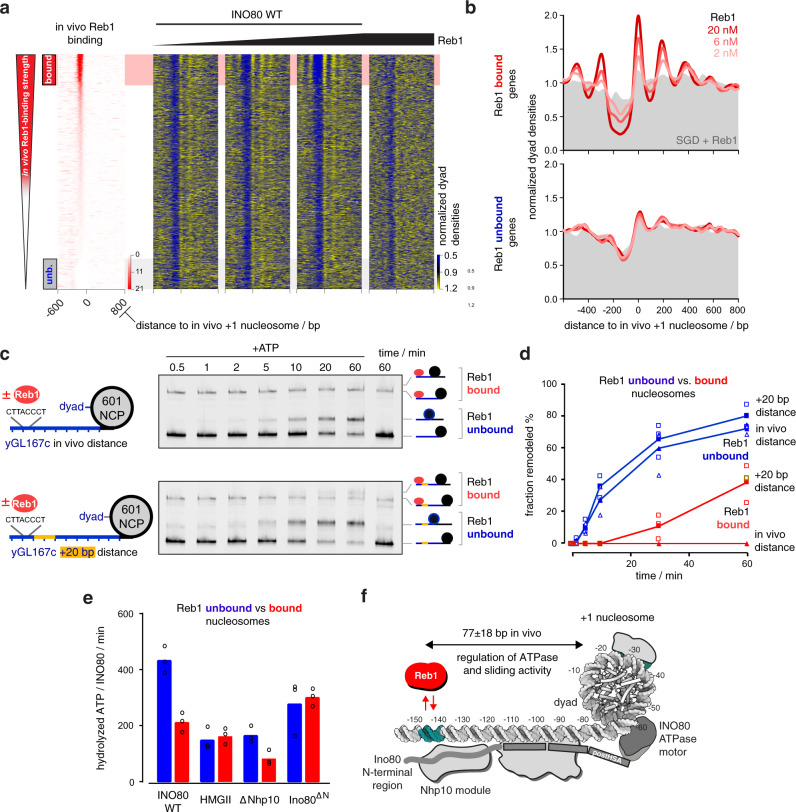
Fig. 6. Reb1 regulates nucleosome positioning by INO80 and INO80’s ATPase and sliding activity.
a Heat maps of MNase-seq data for SGD chromatin assembled with recombinant H. sapiens histones at histone-to-DNA mass ratio 0.4, incubated with recombinant S. cerevisiae WT INO80 and increasing concentrations (ramp denotes 2, 6, and 20 nM) of recombinant Reb1. Right most heat map shows sample prepared with embryonic D. melanogaster histones. Heat maps are aligned at in vivo +1 nucleosome positions and sorted according to decreasing (top to bottom) anti-Reb1 SLIM-ChIP score (in vivo Reb1 binding82) shown in leftmost heat map. Horizontal red or gray shading highlights genes with strong (top 12.5%, “bound”) or weak (bottom 12.5%, “unb.” for unbound) in vivo Reb1 promotor-binding, respectively. Single replicates (replicate 1) were plotted, see Supplementary Fig. 4a and Supplementary Data 1 for all replicates. b Composite plots of MNase-seq data as in a averaged over 620 genes highlighted in red (Reb1 bound, top) or gray (Reb1 unbound, bottom) in a. Gray backgrounds show respective composite plots of SGD chromatin to which Reb1, but not INO80 WT, were added. c Left: mononucleosome substrate design with 80 bp (yGL167c-NCP601, top) or 100 bp (yGL167c-20-NCP601, bottom) linker DNA taken from a promoter (yGL167c) with in vivo-like +1 nucleosome positioning by INO8029 in vitro and strong INO80 binding in vivo41. Guided by its dyad position, we replaced the genomic +1 nucleosome sequence of yGL167c with a 601-nucleosome positioning sequence. Right: representative example of nativePAGE nucleosome sliding assay for mononucleosome species as shown to the right of the gel image and with 90 nM mononucleosome, 45 nM Reb1, 10 nM recombinant S. cerevisiae WT INO80 and 1 mM ATP (denoted as +ATP). 60 min time point was also performed without the addition of ATP. Experiments were independently replicated (n = 3). d Quantification of sliding assays with bound (red) or unbound (blue) Reb1 from c and two other replicates (see Source data). Individual datapoints for independent replicates (n = 3) are shown as empty, average values as filled symbols. e NADH-based ATPase assay (25 nM mononucleosomes, 10 nM recombinant S. cerevisiae WT or mutant INO80 complex, and with (red) or without (blue) 25 nM Reb1). Individual datapoints for biological replicates (n = 3) are shown as circles; bar heights correspond to respective average values. f Structural data44,47 and biochemical mapping48 suggest a putative binding architecture of INO80 which may bridge Reb1 and +1 nucleosomes. Allosteric communication occurs across a distance of more than 70 bp (in vivo median distance of 77 ± 18 bp measured between ChIP-exo mapped Reb1 binding motifs57 and MNase-seq derived +1 nucleosome dyads87; see also accompanying paper39).