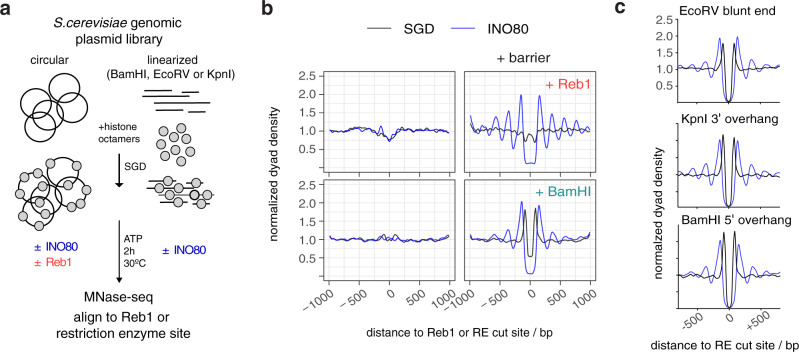
Fig. 8. DNA ends are potent barriers for nucleosome positioning by INO80.
a Overview (analogous to Fig. 2a) of reconstitution with circular versus RE-precleaved plasmid libraries. b Composite plots (merged replicates, n = 2) of MNase-seq data aligned at barrier sites, i.e., anti-Reb1-SLIM-ChIP-defined Reb1 sites or BamHI sites for: top, SGD prepared with circular plasmid library and incubated without (SGD) or with recombinant S. cerevisiae WT INO80 (INO80) and with (+Reb1) or without 20 nM Reb1 as indicated, and bottom: as top but with BamHI-precleaved library if indicated (+BamHI). c As b, but for SGD chromatin with plasmid libraries precleaved with the indicated RE and data aligned at the indicated RE cut sites and only one replicate each (Supplementary Data 1). Strong peaks flanking cut RE sites in SGD chromatin without INO80 remodeling reflected an MNase-seq bias, i.e., due to the pre-cleavage, the probability is increased that MNase releases a mononucleosomal fragment with the cut site as one end relative to releasing fragments from other genomic regions.