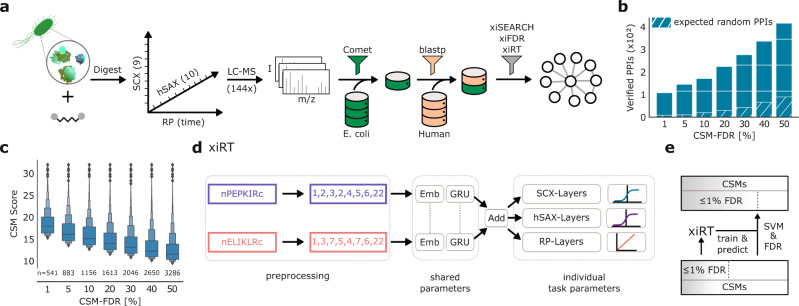
Fig. 1. Workflow overview.
a Experimental and data analysis workflow. The soluble high-molecular-weight proteome of E. coli lysate was crosslinked and the digest sequentially fractionated by strong-cation exchange chromatography (SCX) (9 fractions collected), hydrophilic strong-anion exchange chromatography (hSAX) (10 pools collected), and finally by reversed-phase chromatography (RP) coupled to the MS. The protein database for the crosslink search was created by a linear peptide search with Comet and a sequence-based filter using BLAST. For each E. coli protein in the final database (green) a human protein was added as a control (pale orange). b Potential for false-negative PPI identifications. Verified PPIs are estimated from matches to the STRING/APID databases. PPIs are computed based on CSM-level FDR. Estimated random hits correspond to the average number of semi-randomly drawn pairs (first protein was randomly selected from the STRING/APID database and the second protein was drawn from the FASTA file). Gained PPIs accentuate the additional information that is available in the data at higher FDR. c Decrease of heteromeric CSM scores based on spectral evidence with increasing CSM-FDR. Boxenplot shows the median and 50% of the data in the central boxes while each successive level outward represents half of the remaining data. The sample size for each FDR category is given below the boxes. d xiRT network architecture to predict multi-dimensional retention times. A crosslinked peptide is represented as two individual inputs to xiRT. xiRT uses a Siamese network architecture that shares the weights of the embedding and recurrent layers. Individual layers for the prediction tasks are added with custom activation functions (sigmoid/linear functions for fractionation/regression tasks, respectively). e Rescoring workflow. The predictions from xiRT are combined with xiSCORE’s output to rescore CSMs using a linear support vector machine (SVM), consequently leading to more matches at constant confidence. Source data are provided as a Source Data file.