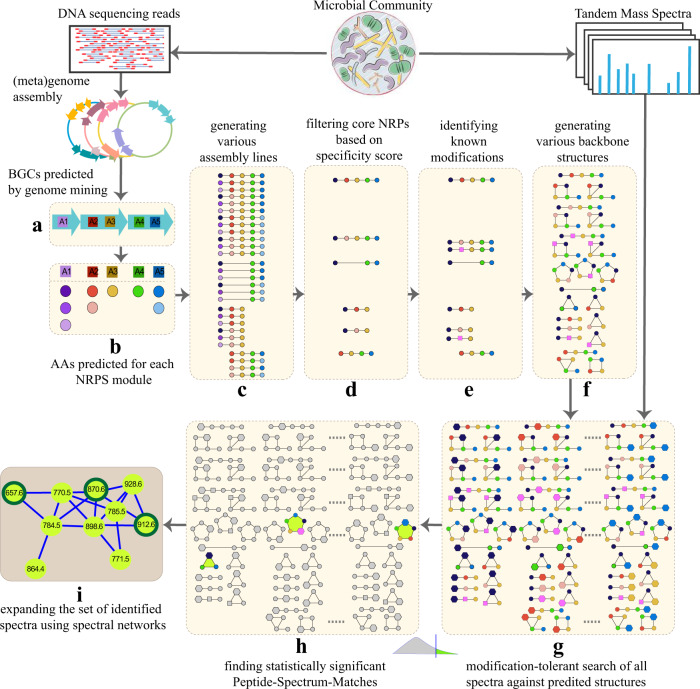
Fig. 1. NRPminer pipeline.
a Predicting NRPS BGCs using antiSMASH16. Each ORF is represented by an arrow, and each A-domain is represented by a square, b predicting putative amino acids for each NRP residue using NRPSpredictor2 (ref. 15), colored circles represents different amino acids (AAs), c generating multiple assembly lines by considering various combinations of ORFs and generating all putative core NRPs for each assembly line in the identified BGC (for brevity only assembly lines generated by deleting a single NRPS unit are shown; in practice, NRPminer considers loss of up to two NRPS units, as well as single and double duplication of each NRPS unit), d filtering the core NRPs based on their specificity scores, e identifying domains corresponding to known modifications and incorporating them in the selected core NRPs (modified amino acids are represented by purple squares), f generating linear, cyclic and branch-cyclic backbone structures for each core NRP, g generating a set of high-scoring PSMs using modification-tolerant VarQuest43 search of spectra against the database of the constructed putative NRP structures. NRPminer considers all possible mature NRPs with up to one PAM (shown as hexagons) in each NRP structure. For brevity some of the structures are not shown. h Computing statistical significance of PSMs and reporting the significant PSMs, and i expanding the set of identified spectra using spectral networks57. Nodes in the spectral network represent spectra and edges connect “similar” spectra (see “Methods”).