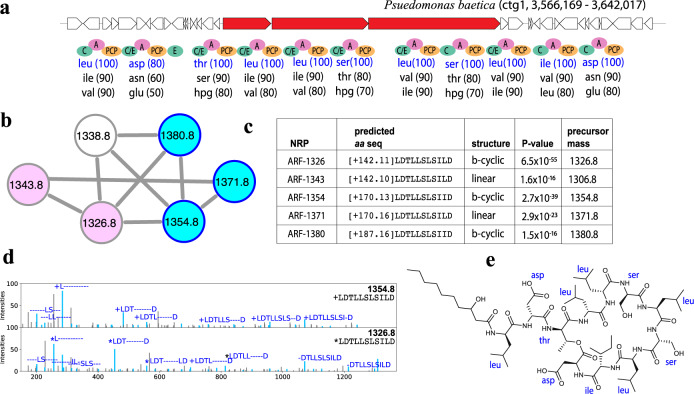
Fig. 8. Arthrofactin (ARF) NRP family.
a The BGCs generating the NRP in Pseudomonas baetica sp. 04-6(1) along with the NRPS genes (shown in red) and A-, C-, C/E-, PCP-, and E-domains in these NRPSs. The rest of the genes in the corresponding contigs are shown in white. Three highest-scoring amino acids for each A-domain in these BGCs (according to NRPSpredictor2 (ref. 15) predictions) are shown below the corresponding A-domains. Amino acids appearing in the known NRP ARF-1354 with amino acid sequence [+170.13]LDTLLSLSILD are shown in blue. b Spectral network formed by the spectra that originate from NRPs in the ARF family. The known arthrofactins are shown in blue, while the purples nodes represent the previously unreported variants identified by NRPminer. All identified athrofactins share the same core NRP LDTLLSLSILD. c Sequences of the identified NRPs in this family (with the lowest p value among all spectra originating from the same NRP). Column “structure” shows if the predicted structure for the identified NRPs is linear or branch-cyclic (shown by b-cyclic). The p values are computed based on MCMC approach using MS-DPR89 with 10,000 simulations. d Two annotated spectra representing the PSMs (with the lowest p values among spectra originating from the same NRPs) corresponding to ARF-1354 and 1326. The two spectra were annotated based on predicted NRPs [+170.13]LDTLLSLSILD (PSM p value 2.7 × 10−39) and [+142.11]LDTLLSLSILD (PSM p value 6.5 10−55), from top to bottom. The “+” and “*” signs represent the addition of [+170.13] and [+142.11], respectively. e The 2D structure of known arthrofactin ARF-1354 (ref. 80). NRPminer identified this NRP with p value 2.7 × 10−39.