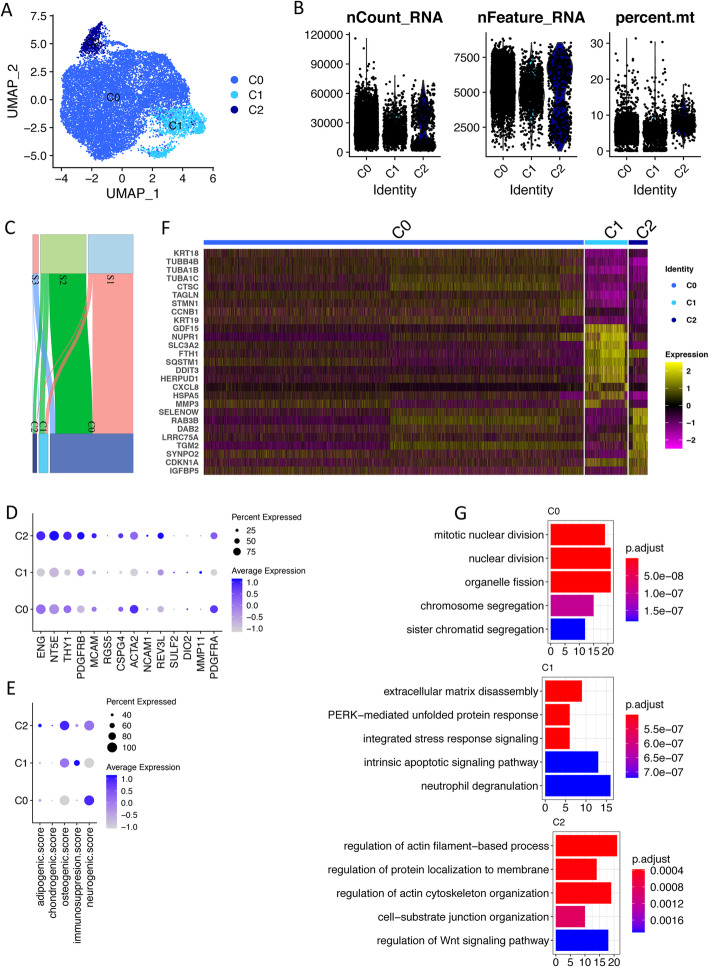
Fig. 4.
scRNA-seq reveals three sub-clusters in cultured secretory ePCs. a Uniform Manifold Approximation and Projection (UMAP) plot of the cellular sub-clusters. Single cells are color coded by cluster annotation. b Violin plot showing the distribution of number of UMIs, number of genes, and percentage of mitochondria reads of single cells in each sub-cluster. c Sankey diagram showing the contribution of cells from each sample to the sub-clusters. d Dot plot showing the expression of MSC markers, perivascular cell markers, smooth muscle cell markers, and stromal fibroblast cell markers across sub-clusters. Circle size indicates the percentage of cells in which the gene expression was detected. Fill color depicts the averaged normalized expression level of all cells within that sub-cluster. e Dot plot showing the score value of adipogenic, chondrogenic, osteogenic, and neurogeneic differentiation and immunomodulation potential across sub-clusters. The score for each cell for each term was firstly calculated by averaging the normalized-expression value of markers in each term. f Heat map of top upregulated genes in sub-clusters C0, C1, and C2. Sub-clusters were clearly separated. g Top significant enriched biological processes from gene ontology analysis based on upregulated genes identified in each sub-cluster. P.adjust, adjusted p value, value on x axis is the enrichment fold