Abstract
Abstract
The progression of prion diseases is accompanied by the accumulation of prions in the brain. Ablation of microglia enhances prion accumulation and accelerates disease progression, suggesting that microglia play a neuroprotective role by clearing prions. However, the mechanisms underlying the phagocytosis and clearance of prion are largely unknown. The macrophage scavenger receptor 1 (Msr1) is an important phagocytic receptor expressed by microglia in the brain and is involved in the uptake and clearance of soluble amyloid-β. We therefore asked whether Msr1 might play a role in prion clearance and assessed the scavenger function of Msr1 in prion pathogenesis. We found that Msr1 expression was upregulated in prion-infected mouse brains. However, Msr1 deficiency did not change prion disease progression or lesion patterns. Prion deposition in Msr1 deficient mice was similar to their wild-type littermates. In addition, prion-induced neuroinflammation was not affected by Msr1 ablation. We conclude that Msr1 does not play a major role in prion pathogenesis.
Key messages
Msr1 expression is upregulated in prion-infected mouse brains at the terminal stage
Msr1 deficiency does not affect prion disease progression
Msr1 does not play a major role in prion clearance or prion pathogenesis
Microglia-mediated phagocytosis and clearance of Aβ and prion may adopt distinct molecular pathways
Supplementary Information
The online version contains supplementary material available at 10.1007/s00109-021-02061-7.
Keywords: Prion disease, Microglia, Macrophage scavenger receptor 1, Phagocytosis, Neuroinflammation
Introduction
Prion diseases are transmissible and fatal neurodegenerative disorders that affect both human and animals. This disease category comprises Creutzfeldt-Jakob disease (CJD), fatal familial insomnia (FFI) and Gerstmann-Sträussler-Scheinker syndrome (GSS) in humans, scrapie in sheep and goat, bovine spongiform encephalopathy (BSE) in cattle, and chronic wasting disease (CWD) in cervids [1]. Prion diseases are thus far incurable. The infectious agent mainly consists of scrapie prion protein (PrPSc), which is a misfolded isoform of the host-encoded cellular prion protein (PrPC). PrPSc acts as a propagon, which can seed a self-perpetuating reaction to recruit and convert PrPC to an aggregated conformation. The deposition of PrPSc in the central nervous system (CNS), together with neuronal loss, spongiform changes (or termed vacuolization), astrogliosis and conspicuous microglial activation, constitutes the characteristic molecular pathology of prion disease [2].
Microglia are the primary innate immune cells and phagocytes of the CNS, exerting a neuroprotective role in prion pathogenesis [3]. We have reported that pharmacogenetic removal of microglia greatly enhances PrPSc accumulation in prion-infected cultured organotypic cerebellar slices (COCS) and in mice [4, 5]. However, the molecular mechanisms underlying prion clearance by microglia are largely unknown [1]. Lack of milk-fat globule EGF factor VIII (Mfge8) was reported to enhance prion pathogenesis in a mouse strain-dependent manner, suggesting that Mfge8 can facilitate engulfment of PrPSc aggregates by microglia [6, 7]. Developmental endothelial locuse1 (Del-1) is a structural and functional homolog of Mfge8 and therefore was a further candidate modifier of prion removal. However, Del-1 deficiency neither changed prion deposition nor prion pathogenesis in mice, suggesting that Del-1 does not complement Mfge8 in prion clearance [8]. Also triggering receptor expressed on myeloid cells 2 (TREM2), a phagocytic receptor expressed on microglia, modulates prion-induced microglial activation but does not contribute to prion clearance [9]. Hence, the molecules that are involved in prion clearance are complex and require further study.
Microglia express various receptors that facilitate sensing and phagocytosis of pathogens and misfolded protein aggregates; these include toll-like receptors (TLRs), scavenger receptors(SRs), Fc receptors, complement receptors, triggering receptor expressed on myeloid cells-2 (TREM2), myeloid cell surface antigen CD33, and others [10]. Importantly, variants of the TREM2 and CD33 genes are risk factors for Alzheimer’s disease (AD) [11–13], probably due to impaired uptake and clearance of amyloid-β (Aβ) [14, 15]. Although TREM2 is not a main transducer of prion clearance [9], the role of other microglial receptors in prion pathogenesis merits further investigations.
Msr1, also known as scavenger receptor a1 (Scara1), is a type II transmembrane glycoprotein mainly expressed by microglia in CNS [16]. Msr1 has been involved in many macrophage-associated physiological and pathological conditions such as neurodegenerative diseases [17]. As an important phagocytic receptor, Msr1 can mediate uptake of fibrillary amyloid β (Aβ) in vitro [18–20]. Msr1 deficiency in a mouse model of AD markedly accelerates Aβ accumulation and disease progression, whereas pharmacological upregulation of Msr1 leads to enhanced Aβ clearance. These results collectively suggest that Msr1 is essential for clearing soluble Aβ [21, 22]. Since both Aβ and prion are extracellular misfolded protein aggregates, they may share similar molecular pathways by which microglia take up and degrade the protein aggregates.
In this study, we aimed to investigate whether Msr1 may have a similar function in prion clearance and pathogenesis. We first determined the Msr1 expression in a mouse model of prion disease and found Msr1 expression was significantly increased in prion-infected mouse brain. After prion inoculation, we then observed that Msr1-/- mice showed disease progression similar to their hemizygous (Msr1+/-) and wild-type (Msr1+/+) littermates. Besides, prion deposition and seeding dose were not altered by Msr1 deficiency, suggesting that Msr1 is not involved in prion clearance. Furthermore, Msr1 deficiency did not affect prion-induced neuroinflammation. We therefor conclude that Msr1 is not a major player in prion clearance and does not influence prion pathogenesis.
Material and methods
Ethical statement
All animal experiments were carried out in strict accordance with the Rules and Regulations for the Protection of Animal Rights (Tierschutzgesetz and Tierschutzverordnung) of the Swiss Bundesamt für Lebensmittelsicherheit und Veterinärwesen and were pre-emptively approved by the Animal Welfare Committee of the Canton of Zürich (permit # 41/2012).
Animals
Msr1-/- mice were generated by inserting a neomycin cassette into the EcoRI site in exon 4, which encodes the alpha helical coiled-coil structure essential for the formation of functional trimeric receptors ([23]; JAX stock #006096). Msr1-/- mice were first backcrossed to C57BL/6 J mice to obtain Msr1+/- offspring, which were then intercrossed to generate Msr1+/+ (wild type), Msr1+/- and Msr1-/- mice for experiments described here. All animals were maintained in high hygienic grade facility under a 12 h light/12 h dark cycle (from 7 am to 7 pm) at 21±1 °C and fed with diet and water ad libitum.
Prion inoculation
Mice were intracerebrally (i.c) inoculated with 30 μl of brain homogenate diluted in PBS with 5% BSA and containing 3 × 105 LD50 units of the Rocky Mountain Laboratories scrapie strain (passage 6, thus called RML6). Mice were monitored and actions were taken to minimize animal suffering and distress according to details described previously [24]. Scrapie was diagnosed according to clinical criteria (ataxia, limb weakness, front leg paresis and rolling). Mice were sacrificed by CO2 inhalation on the day of appearance of terminal clinical signs of scrapie (specific criteria referred to [24]), organs were taken and then were either snap-frozen for biochemical analysis or fixed in 4% formalin for histological assessment. The time elapsed from prion inoculation to the terminal stage of disease was defined as incubation time for the survival study.
Quantitative real-time PCR (qRT-PCR)
Total RNA from was extracted using TRIzol (Invitrogen Life Technologies) according to the manufacturer’s instruction. The quality of RNA was analyzed by Bioanalyzer 2100 (Agilent Technologies), RNAs with RIN > 8 were used for cDNA synthesis. cDNAs were synthesized from ~ 1 μg total RNA using QuantiTect Reverse Transcription kit (QIAGEN) according to the manufacturer’s instruction. Quantitative real-time PCR (qRT-PCR) was performed using the SYBR Green PCR Master Mix (Roche) on a ViiA7 Real-Time PCR system (Applied Biosystems). The following primer pairs were used: GAPDH sense 5´-CCA CCC CAG CAA GGA GAC T-3´; antisense, 5´-GAA ATT GTG AGG GAG ATG CT-3´. Msr1 sense 5´- TGA ACG AGA GGA TGC TGA CTG -3´; antisense, 5´- GGA GGG GCC ATT TTT AGT GC -3´. TNFα sense, 5´-CAT CTT CTC AAA ATT CGA GTG ACA A-3´; antisense, 5´-TGG GAG TAG ACA AGG TAC AAC CC-3´. IL-1β sense, 5´-CAA CCA ACA AGT GAT ATT CTC CAT G-3´; antisense, 5´-GAT CCA CAC TCT CCA GCT GCA-3´. IL-6 sense, 5´-TCC AAT GCT CTC CTA ACA GAT AAG-3´; antisense, 5´-CAA GAT GAA TTG GAT GGT CTT G -3´. Expression levels were normalized using GAPDH.
Immunohistochemistry
Prion-infected brain tissues were harvested and fixed in formalin, followed by treatment with concentrated formic acid for 60 min to inactivate prion infectivity and embedded in paraffin. Paraffin sections (2 μm) of brains were stained with hematoxylin/eosin (HE) to visualize prion-induced lesions and vacuolation. For the histological detection of partially proteinase K-resistant prion protein deposition, deparaffinized sections were pretreated with formaldehyde for 30 min and 98% formic acid for 6 min, and then washed in distilled water for 30 min. Sections were incubated in Ventana buffer and stains were performed on a NEXEX immunohistochemistry robot (Ventana instruments, Switzerland) using an IVIEW DAB Detection Kit (Ventana). After incubation with protease 1 (Ventana) for 16 min, sections were incubated with anti-PrP SAF-84 (1:200, SPI bio, A03208) for 32 min. Sections were counterstained with hematoxylin. To detect astrogliosis and microglial activation, brain sections were deparaffinized through graded alcohols, anti-GFAP antibody (1:300; DAKO, Carpinteria, CA) were applied for astrogliosis, and anti-AIF1 antibody (1:1000; Wako Chemicals GmbH, Germany) was used for highlighting activated microglial cells. Stainings were visualized using DAB (Sigma-Aldrich), and hematoxylin counterstain was subsequently applied. Sections were imaged using a Zeiss Axiophot light microscope. Quantification of SAF84 staining was performed on acquired images. Regions of interest were drawn, and the average signal density was quantified using Image J software (National Institutes of Health).
Western blot analysis
To detect PrPC in the mouse brains, one hemisphere of each brain was homogenized with RIPA buffer. Total protein concentration was determined using the bicinchoninic acid assay (Pierce). ~8 μg proteins were loaded and separated on a 12% Bis-Tris polyacrylamide gel (NuPAGE, Invitrogen) and then blotted onto a nitrocellulose membrane. Membranes were blocked with 5% wt/vol Topblock (LuBioScience) in PBS supplemented with 0.1% Tween 20 (vol/vol) and incubated with primary antibodies POM1 in 1% Topblock (400 ng ml−1) overnight. After washing, the membranes were then incubated with secondary antibody horseradish peroxidase (HRP)-conjugated goat anti–mouse IgG (1:10,000, Jackson ImmunoResearch, 115-035-003). Blots were developed using Luminata Crescendo Western HRP substrate (Millipore) and visualized using the Stella system (model 3000, Bio-Rad). To avoid variation in loading, the same blots were stripped and incubated with anti-actin antibody (1:10,000, Millipore). The PrPC signals were normalized to actin as a loading control.
To detect PrPSc, prion infected forebrains were homogenized in sterile 0.32 M sucrose in PBS. Total protein concentration was determined using the bicinchoninic acid assay (Pierce). Samples were adjusted to 20 μg protein in 20 μl and digested with 25 μg ml−1 proteinase K for 30 min at 37 °C. PK digestion was stopped by adding loading buffer (Invitrogen) and boiling samples at 95 °C for 5 min. Proteins were then separated on a 12% Bis-Tris polyacrylamide gel (NuPAGE, Invitrogen) and blotted onto a nitrocellulose membrane. POM1 and horseradish peroxidase (HRP)-conjugated goat anti–mouse IgG were used as primary and secondary antibodies, respectively. Blots were developed using Luminata Crescendo Western HRP substrate (Millipore) and visualized using the FUJIFILM LAS-3000 system. To detect GFAP and AIF1 in prion-infected brains by Western blot, 20 μg of total brain protein were loaded and anti-GFAP antibody (D1F4Q) XP Rabbit mAb (1:3000; Cell Signaling Technology, 12389), anti-AIF1 antibody (1:1000; Wako Chemicals GmbH, Germany, 019-19741) and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:10,000, Jackson ImmunoResearch, 111-035-045) were used as primary and secondary antibodies, respectively. Actin was used as the loading control.
Real-time quaking induced conversion assay (RT-QuIC)
RT-QuIC assays of prion-infected mouse brain homogenates were performed as previously described [8, 25]. Briefly, recombinant hamster full-length (23–231) PrP was expressed in Rosetta2(DE3)pLysS E. coli competent cells and purified by affinity chromatography using Ni2+-nitrilotriacetic acid Superflow resin (QIAGEN). In the RT-QuIC assay, recombinant HaPrP was used as substrate for PrPSc-catalyzed conversion. RT-QuIC reactions containing HaPrP substrate protein at a final concentration of 0.1 mg mL-1 in PBS (pH 7.4), 170 mM NaCl, 10 μM EDTA, and 10 μM Thioflavin T were seeded with 2 μL of serially diluted brain homogenates in a total reaction volume of 100 μL. NBH- and RML6-treated brain homogenates were used as negative and positive controls, respectively. The RT-QuIC reactions were amplified at 42 °C for 100 h with intermittent shaking cycles of 90 s shaking at 900 rpm in double orbital mode and 30 s resting using a FLUOstar Omega microplate reader (BMG Labtech). Aggregate formation was followed by measuring the thioflavin T fluorescence every 15 min (450 nm excitation, 480 nm emission; bottom read mode).
Statistical analysis
Results are presented as the mean of replicas ± standard error of the mean (SEM). Statistical significance between experimental groups was assessed using an unpaired Student’s t test. For prion inoculation experiments, incubation times were analyzed using the Kaplan-Meier method and compared between groups using Log-rank (Mantel-Cox) test. p values < 0.05 were considered statistically significant.
Results
Prion infection upregulates Msr1 expression in the mouse brain
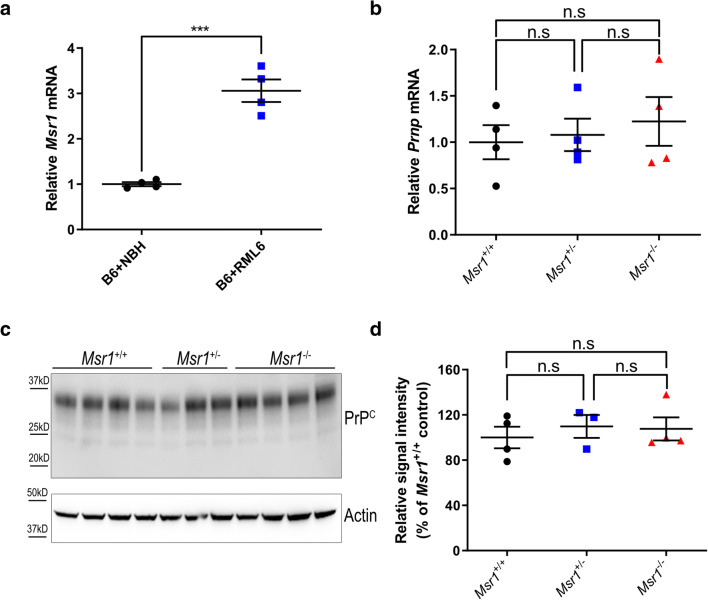
Expression of Msr1 could be upregulated by various forms of brain injury and neurodegeneration [16, 26–29]. To test whether prion infection affects Msr1 expression in the mouse brain, we intracerebrally (i.c) inoculated 30 μl of diluted CD1 mouse brain homogenate containing 3 × 105 LD50 (50% lethal dose) units of RML6 (a prion strain originated from the Rocky Mountain Laboratory, serially passaged to No. 6, hence termed RML6) into C57BL/6 J mice. C57BL/6 J littermates inoculated with noninfectious brain homogenate (NBH) were used as control. Prion-inoculated mice were euthanized and brains were collected when they showed severe scrapie sign and reached the terminal stage of disease, NBH-inoculated control mice were sacrificed after the same incubation time. Since we could not commercially obtain a sensitive and specific antibody detecting the Msr1 protein in mouse brains (supplemental Fig. 1), we assessed Msr1 expression levels using quantitative reverse-transcription PCR (qRT-PCR). The results showed that Msr1 expression was significantly enhanced in prion-inoculated mouse brain, compared to NBH inoculated mouse (Fig. 1a), suggesting that Msr1 expression can be efficiently upregulated by prion infection.
Fig. 1.
(a) qRT-PCR of Msr1 expression in mouse brains. Msr1 expression was significantly increased in RML inoculated mouse brain, compared to NBH inoculated mouse brain (n = 4, p < 0.001). (b) Prnp qRT-PCR in Msr1+/+ (WT), Msr1+/-, and Msr1-/- mouse brains (n = 4, n.s p > 0.05). No significant difference of Prnp mRNA was observed between three groups. c and d PrPC Western blot (c) and densitometric quantification of the Western blot (d) in Msr1+/+ (WT), Msr1+/-, and Msr1-/- mouse brains (n=3~4, n.s p>0.05). No significant difference of PrPC protein was observed between three groups. Abbreviations: Msr1, macrophage scavenger receptor 1 gene; Prnp, murine prion gene; PrPC, cellular prion protein; qRT-PCR, quantitative real-time polymerase chain reaction; RML6, Rocky Mountain Laboratories scrapie strain, passage 6; NBH, noninfectious brain homogenates; WT, wild type
Consistent with our qRT-PCR results, a time course study of hippocampal transcriptome of prion-infected mice by RNA sequencing (RNA-Seq) showed that Msr1 mRNA level was significantly increased at the terminal stage of RML-inoculated mice, but not in NBH-inoculated control mice (supplemental Fig. 2a). Interestingly, however, during the preclinical stage and even at the early stage after onset of clinical symptoms, Msr1 mRNA levels were not significantly changed (supplemental Fig. 2a). Considering that Msr1 is mainly expressed by microglia in the CNS, and the microglia can be conspicuously activated and proliferate upon prion infection, we aimed to determine whether the upregulation of Msr1 mRNA is due to the increased microglia number. We found that mRNA levels of various microglia markers including Aif1, CD68, and Itgax were almost simultaneously induced in prion-inoculated mice from 16 weeks post-inoculation on (supplemental Figs. 2b–d), when Msr1 mRNA was not significantly altered yet [30]. Moreover, a cell-type-specific ribosomal profiling using CX3CR1-CreER mice revealed that Msr1 was indeed upregulated in microglia only at the terminal stage (supplemental Figure 2E) [31]. Therefore, these results suggest that upregulation of Msr1 expression was not merely an effect of increased microglia number after prion infection, but displayed unique pattern and dynamics.
PrPC expression levels in mouse brains are the major determinants of the susceptibility to prion diseases and progression rate of the diseases [32, 33]. To assess whether Msr1 deficiency could influence PrPC expression, we performed qRT-PCR and Western blot to measure Prnp mRNA transcripts and PrPC protein levels in Msr1+/+, Msr1+/-, and Msr1-/- littermates. The results showed that Msr1 deficiency altered neither the Prnp mRNA (Fig. 1b) nor the protein levels of PrPC (Figs. 1c–d, uncropped Western blots are shown in Supplementary Fig. 3a–b) in mouse brains.
Prion disease progression, lesion pattern, or PrPSc accumulation are not altered by Msr1 deficiency
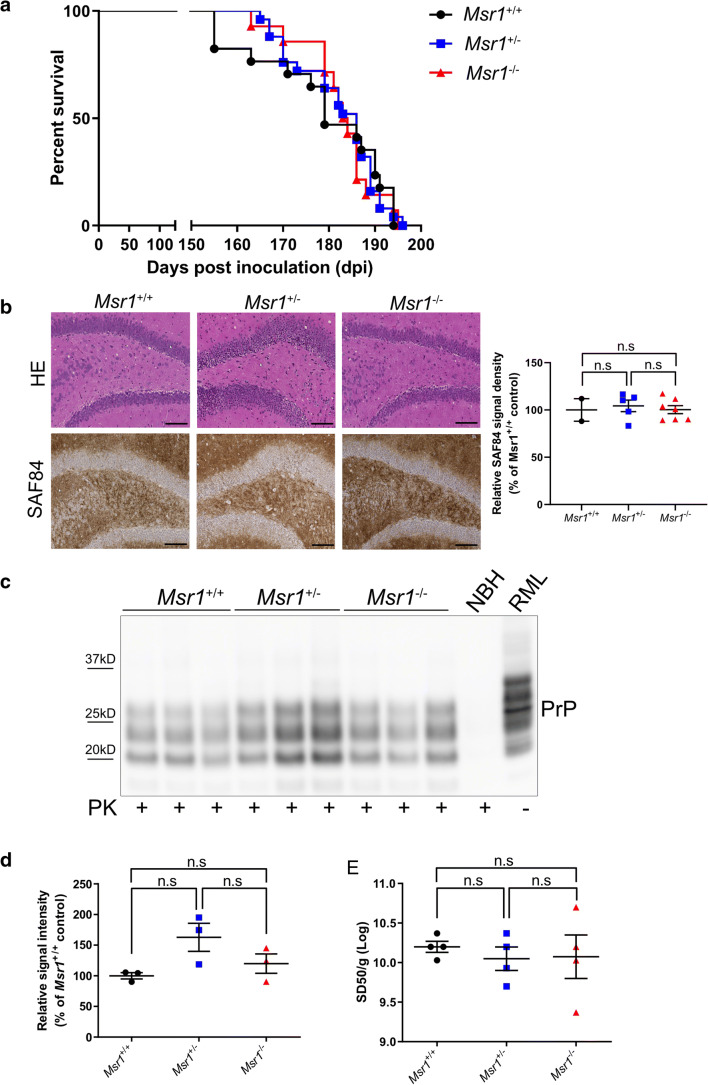
To assess the function of Msr1 in prion pathogenesis, we next tested whether Msr1 ablation could affect prion disease progression and alter prion-mediated lesion pattern in mouse brains. We intracerebrally (i.c) inoculated 30 μl of RML6 prions into Msr1+/+, Msr1+/-, and Msr1-/- littermates. RML6-inoculated mice were checked and monitored every other day for scrapie symptoms. Mice were euthanized and brains were collected when they showed severe scrapie sign and reached the terminal stage of disease. Incubation times were calculated as the time from initial prion inoculation until terminal disease stage. We observed that all Msr1+/+, Msr1+/-, and Msr1-/- mice succumbed to prion disease at a similar progression rate (median survival: 179 dpi for Msr1+/+ mice (n = 17), 186 dpi for Msr1+/-(n = 25) and 183.5 dpi for Msr1-/- mice (n = 14), p = 0.99) (Fig. 2a). The above results indicate that Msr1 ablation does not overtly influence progression of prion disease.
Fig. 2.
(a) Survival curve of Msr1+/+ (WT), Msr1+/- and Msr1-/- littermates intracerebrally inoculated with RML6. There was no significant difference between three groups in both genders (n = 14–25, n.s p > 0.05). (b) Left: Representative histology of terminally sick mouse brains from Msr1+/+ (WT), Msr1+/- and Msr1-/- littermates stained for H&E and SAF84. Right: quantification of SAF84 staining. There was no obvious difference between three groups in vacuolation, lesion pattern and PrPSc deposition in hippocampus (n = 2–7, n.s p > 0.05). c and d Western blot for proteinase K resistant PrPSc in terminally sick mouse brains (c) and densitometric quantification of the Western blot (d). There was no significantly difference between Msr1+/+ (WT), Msr1+/- and Msr1-/- littermates (n=3, n.s p > 0.05). (E) RT-QuIC assay of terminally sick mouse brains from Msr1+/+ (WT), Msr1+/- and Msr1-/- littermates showed similar level of 50% prion seeding dose in these three groups (n = 4, n.s p > 0.05). Abbreviations: Msr1, macrophage scavenger receptor 1; PrPSc, scrapie-associated prion protein; RML6, Rocky Mountain Laboratories scrapie strain, passage 6; NBH, noninfectious brain homogenates; WT, wild type; RT-QuIC, real-time quaking-induced conversion assay
We then analyzed and compared the histology of brain sections collected and prepared from RML6-inoculated terminally sick Msr1+/+, Msr1+/- and Msr1-/- mice. The classical histological characteristics of prion disease, especially the spongiform changes (or vacuolation), were observed in all mice in different groups. Lesion pattern analysis also failed to show any qualitative distinctions between different genotypes (Figure 2B). These results suggest that lack of Msr1 does not obviously affect prion-caused lesion profile in mouse brains.
If Msr1 contributed to prion clearance as it does to Aβ, Msr1-/- mice would accumulate more PrPSc deposits in their brains. We therefore first performed PrPSc staining on brain sections prepared from RML6-inoculated terminally sick Msr1+/+, Msr1+/- and Msr1-/- mice. Unexpectedly, we observed a similar PrPSc deposition level and pattern in mouse brains with the different genotypes (Fig. 2b). We next performed Western blot to detect and assess proteinase K (PK)-resistant PrPSc levels in brains of terminally sick mice. We again found a similar PrPSc accumulation level in Msr1+/+, Msr1+/-, and Msr1-/- mouse brains (Figs. 2c–d, uncropped Western blots are shown in Supplementary Fig. 3c). Additionally, we measured the prion seeding activity in brain homogenates of RML6-inoculated terminally sick Msr1+/+, Msr1+/-, and Msr1-/- mice by real time-quaking induced conversion assay (RT-QuIC). We detected an undistinguishable 50% seeding dose (SD50) of prion in all three groups (Fig. 2e). Taken together, these results indicate that Msr1 does not play a major role in prion clearance or PrPSc accumulation in mouse brains.
Prion-induced neuroinflammation is not affected by Msr1 deficiency
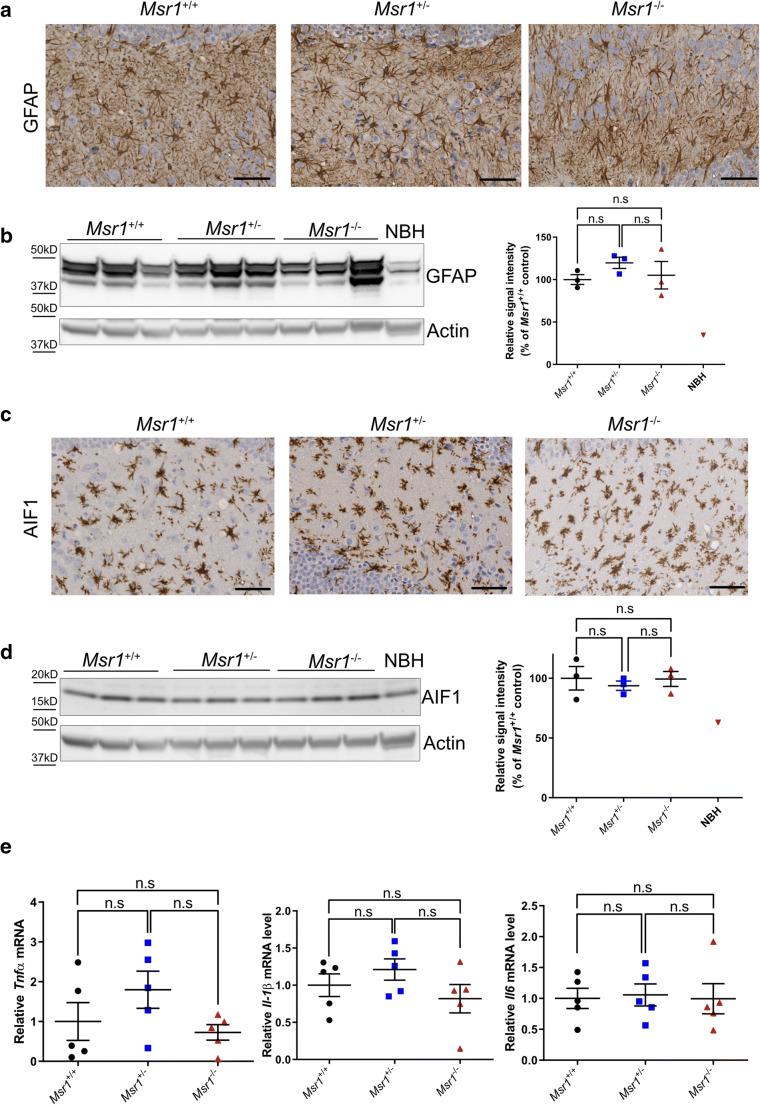
Msr1 is a scavenger receptor that could modulate immune response in CNS [34–36]. To test whether Msr1 exhibits an anti-inflammatory function in prion pathogenesis, we analyzed and compared astrogliosis and microglial activation in prion-infected terminally sick Msr1+/+, Msr1+/- and Msr1-/- mice. First, histology and Western blots did not identify obvious differences in GFAP (glial fibrillary acidic protein, an astrocytic marker) level between the three groups with different genotypes (Figs. 3a–b, uncropped Western blots are shown in Supplementary Figs. 3d–e). We next performed histological examinations and Western blots to detect AIF1 (allograft inflammatory factor-1, a microglial marker) in RML6-inoculated terminally sick Msr1+/+, Msr1+/-, and Msr1-/- mouse brains. We again observed similar levels of AIF1 between the three groups (Figs. 3c–d, uncropped Western blots are shown in Supplementary Figs. 3f–g). These results together suggest that Msr1 deficiency does not significantly affect prion-induced astrogliosis or microglial activation.
Fig. 3.
a Representative immunohistochemical staining for GFAP in the hippocampus of terminally sick mouse brains from Msr1+/+ (WT), Msr1+/-, and Msr1-/- littermates. b Left: Western blot for GFAP in terminally sick mouse brains. Right: densitometric quantification of the Western blot showed no significant difference of GFAP levels between Msr1+/+ (WT), Msr1+/- and Msr1-/- littermates (n =3–4, n.s p > 0.05). c Representative immunohistochemical staining for AIF1 in the hippocampus of terminally sick mouse brains from Msr1+/+ (WT), Msr1+/- and Msr1-/- littermates. d Left: Western blot for AIF1 in terminally sick mouse brains. Right: densitometric quantification of the Western blot showed no significant difference of AIF1 levels between Msr1+/+ (WT), Msr1+/-, and Msr1-/- littermates (n =3–4, n.s p > 0.05). e qRT-PCR of cytokines TNFα, IL-1β and IL-6 expression revealed similar mRNA levels of these cytokines (Tnfα, Il1β,Il6) in terminally sick Msr1+/+ (WT), Msr1+/-, and Msr1-/- littermates (n = 5, n.s p > 0.05). Abbreviations: Msr1, macrophage scavenger receptor 1; GFAP, glial fibrillary acidic protein; AIF1, Ionized calcium binding adaptor molecule 1; WT, wild type; qRT-PCR, quantitative real-time polymerase chain reaction
We next performed qRT-PCR to analyze the expression levels of proinflammatory cytokines (TNFα, IL-1β and IL6) in RML6-inoculated terminally sick Msr1+/+, Msr1+/-, and Msr1-/- mouse brains. Again, this experiment failed to detect any overt differences in cytokine expression levels between the three groups (Fig. 3E), suggesting that Msr1 ablation does not change the cytokine profiles of prion-infected mouse brains.
Discussion
It is becoming increasingly clear that neuroinflammation, characterized by astrogliosis and microglial activation, is a hallmark of many neurodegenerative diseases including prion diseases [37]. Emerging evidence from multiple genome-wide association studies (GWAS) suggests that microglia may play a central role in neurodegenerative diseases because most identified risk factors are expressed by microglia [11, 12, 38–42]. The molecular mechanisms by which microglia contributes to the pathogenesis of neurodegenerative diseases are the subject of intense investigations. Insight into these questions will enable not only the understanding of disease pathogenesis but also the development of novel therapeutic strategies combating these disorders [43].
As the primary tissue-resident macrophages in the CNS, microglia exert diverse functions in the brain. Microglia play important roles during brain development by pruning neuronal synapses [44]. In adult brain, microglia could modulate learning and memory [45] and constantly survey their local milieu for signals of danger and injury, thus serving as important sensors and defenders upon challenges [46, 47]. Under pathological conditions such as in prion disease, microglia are found to play an overall neuroprotective role by clearing prions. Depletion or deficiency of microglia results in impaired prion clearance, enhanced PrPSc deposition, and deteriorated prion pathogenesis [5]. These findings collectively highlight the need to better understand the molecular mechanisms underlying microglia-mediated prion clearance [3].
Msr1, a type II transmembrane glycoprotein expressed mainly by microglia in the brain [16], is a member of scavenger receptors that mediate endocytosis of a wide range of ligands including low-density lipoproteins, bacterial pathogens and dead cells [48, 49]. The role of Msr1 in neurodegeneration such as Alzheimer’s disease (AD) was first highlighted in an in vitro study showing that it could facilitate adhesion of microglia to fibrillar Aβ [18]. This finding was further validated by a short hairpin RNA (shRNA) library screen [21]. In mouse models of AD, Msr1 deficiency impairs clearance of soluble Aβ and results in increased Aβ deposition and early mortality, whereas pharmacological upregulation of Msr1 leads to enhanced Aβ clearance [21, 22]. Therefore, it is reasonable to speculate that Msr1 could play a scavenger function for a broad range of misfolded protein aggregates including prions.
In this study, we tested whether Msr1 plays a scavenger function in prion pathogenesis, similar to that in mouse models of AD. Firstly, in contrast to what was observed in aged AD mouse models [50, 51], Msr1 expression was found to be upregulated at the terminal stage upon prion infection. After prion inoculation, we observed that Msr1-/- mice experienced a similar incubation time compared with Msr1+/- and wild-type littermates, suggesting that Msr1 deficiency does not affect prion disease progression. Notably, the PrPSc level and the prion seeding activity were not detectably affected by Msr1 deficiency, indicating that Msr1 is not a major contributor to prion clearance. Moreover, prion-induced neuroinflammation including astrocytosis and microglial activation was not altered in Msr1-/- mouse brains, suggesting that Msr1 deficiency does not overtly affect prion-induced neuroinflammation. Together, these results indicate that Msr1 functions differently in AD and prion diseases.
Limitations of the current study
In the present study, we found that Msr1 expression was upregulated in terminally sick prion-infected mouse brains detected by both qRT-PCR and RNA-seq. Longitudinal study showed that upregulation of Msr1 was not merely due to increased microglia number. Although both analyses were based on brain tissue containing a mixture of various cell types, a cell-type specific ribosomal profiling again found that Msr1 was significantly upregulated in microglia only at the terminal stage. Orthogonal methodologies, such as cell sorting followed by RNA-seq, would be required to confirm the microglia-specific Msr1 expression pattern along the prion progression. Nevertheless, the late upregulation of Msr1 may explain that Msr1 have little impact on the prion pathogenesis. The varied temporal pattern and magnitude of changes of Msr1 expression between prion disease and AD may explain its differential functions in these diseases.
We performed prion infection experiments with an appropriate number of animals (n=14~25 mice per group) and conclude, with a high degree of confidence, that Msr1 deficiency did not overtly affect progression of prion disease. We randomly divided the samples into subgroups for either biochemical analysis or histology encompassing a relatively small number of samples (n = 3–5 mice per group). Because of the increased stochasticity inherent to such subgroups, we observed some variations in these analyses. Notwithstanding these limitations, our results point towards the same direction and are congruent with our conclusion that Msr1 does not play a major role in prion pathogenesis.
Conclusion
Collectively, these results indicate that Msr1 does not play a major role in prion pathogenesis. Together with the discrepant observations of TREM2 in AD and prion disease[9, 14], this study suggests that microglia-mediated phagocytosis and clearance of Aβ and prion may adopt distinct molecular pathways. Further studies are needed to investigate molecular mechanisms underlying microglial uptake and clearance of prions.
Supplementary Information
Western blots using commercial anti-Msr1 antibodies that could not detect specific Msr1 band on brains collected from wild type mice. Msr1-/- mouse brains were used as negative control. (PNG 512 kb)
RNA-Seq data of Msr1 (A) and microglia markers (Aif1, Cd68 and Itgax) (B-D) expression in hippocampi at different time points after prion inoculation. (n=3, except n=2 at 20wpi for NBH, *P<0.05; ***P<0.001). (E) Ribosomal profiling of Msr1 expression in CX3CR1-positive microglia (n=3, except n=2 at terminal stage, ****P<0.0001). Abbreviations: Msr1, macrophage scavenger receptor 1 gene; Aif1, Ionized calcium binding adaptor molecule 1 gene; Cd68, cluster of differentiation 68 gene; Itgax, integrin subunit alpha x gene; RML6, Rocky Mountain Laboratories scrapie strain, passage 6; NBH, noninfectious brain homogenates. (PNG 451 kb)
Full images of the cropped Western blots in Figure 1C (A-B), 2C (C), 3B (D-E) and 3D (F-G). (PNG 802 kb)
Acknowledgments
We thank the teams of the Institute of Neuropathology, University Hospital Zurich and Department of Neurobiology, School of Basic Medical Sciences of Fudan University for technical assistance. The authors thank Elisabeth J. Rushing for reading and editing the article. A. Aguzzi is the recipient of an Advanced Grant of the European Research Council (ERC, No. 670958) and is supported by grants from the European Union (PRIORITY, NEURINOX), the Swiss National Foundation (SNF, including a Sinergia grant), the Swiss Initiative in Systems Biology, SystemsX.ch (PrionX, SynucleiX), and a Distinguished Scientist Award of the Nomis Foundation. C. Zhu is sponsored by Research Startup Funds of Fudan University, Shanghai Pujiang Program (No. 20PJ1401100) and National Natural Science Foundation of China (No. 82071436). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author’s contributions
C. Zhu and A. Aguzzi conceived the project and designed the experiments; B. Li, M. Chen, and C. Zhu conducted the experiments and acquired data. All authors contributed to data analysis.
Funding
Open Access funding provided by Universität Zürich. A. Aguzzi is the recipient of an Advanced Grant of the European Research Council (ERC, No. 670958) and is supported by grants from the European Union (PRIORITY, NEURINOX), the Swiss National Foundation (SNF, including a Sinergia grant), the Swiss Initiative in Systems Biology, SystemsX.ch (PrionX, SynucleiX), and a Distinguished Scientist Award of the Nomis Foundation. C. Zhu is sponsored by Research Startup Funds of Fudan University, Shanghai Pujiang Program (No. 20PJ1401100) and National Natural Science Foundation of China (No. 82071436). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and material
The authors confirm that the data and material supporting the findings of this study are available within the article and its supplementary materials.
Code availability
Not applicable.
Declarations
Ethics approval
All animal experiments were carried out in strict accordance with the Rules and Regulations for the Protection of Animal Rights (Tierschutzgesetz and Tierschutzverordnung) of the Swiss Bundesamt für Lebensmittelsicherheit und Veterinärwesen and were preemptively approved by the Animal Welfare Committee of the Canton of Zürich (permit # 41/2012).
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Adriano Aguzzi, Email: Adriano.Aguzzi@usz.ch.
Caihong Zhu, Email: Caihong_Zhu@fudan.edu.cn.
References
- 1.Aguzzi A, Nuvolone M, Zhu C. The immunobiology of prion diseases. Nat Rev Immunol. 2013;13:888–902. doi: 10.1038/nri3553. [DOI] [PubMed] [Google Scholar]
- 2.Aguzzi A, Zhu C. Five questions on prion diseases. PLoS Pathog. 2012;8:e1002651. doi: 10.1371/journal.ppat.1002651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguzzi A, Zhu C. Microglia in prion diseases. J Clin Invest. 2017;127:3230–3239. doi: 10.1172/JCI90605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falsig J, Julius C, Margalith I, Schwarz P, Heppner FL, Aguzzi A. A versatile prion replication assay in organotypic brain slices. Nat Neurosci. 2008;11:109–117. doi: 10.1038/nn2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu C, Herrmann US, Falsig J, Abakumova I, Nuvolone M, Schwarz P, Frauenknecht K, Rushing EJ, Aguzzi A. A neuroprotective role for microglia in prion diseases. J Exp Med. 2016;213:1047–1059. doi: 10.1084/jem.20151000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kranich J, Krautler NJ, Heinen E, Polymenidou M, Bridel C, Schildknecht A, Huber C, Kosco-Vilbois MH, Zinkernagel R, Miele G, Aguzzi A. Follicular dendritic cells control engulfment of apoptotic bodies by secreting Mfge8. J Exp Med. 2008;205:1293–1302. doi: 10.1084/jem.20071019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kranich J, Krautler NJ, Falsig J, Ballmer B, Li S, Hutter G, Schwarz P, Moos R, Julius C, Miele G, Aguzzi A. Engulfment of cerebral apoptotic bodies controls the course of prion disease in a mouse strain-dependent manner. J Exp Med. 2010;207:2271–2281. doi: 10.1084/jem.20092401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, Li Z, Li B, Pfammatter M, Hornemann S, Aguzzi A. Unaltered prion disease in mice lacking developmental endothelial locus-1. Neurobiol Aging. 2019;76:208–213. doi: 10.1016/j.neurobiolaging.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhu C, Herrmann US, Li B, Abakumova I, Moos R, Schwarz P, Rushing EJ, Colonna M, Aguzzi A. Triggering receptor expressed on myeloid cells-2 is involved in prion-induced microglial activation but does not contribute to prion pathogenesis in mouse brains. Neurobiol Aging. 2015;36:1994–2003. doi: 10.1016/j.neurobiolaging.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Doens D, Fernandez PL. Microglia receptors and their implications in the response to amyloid beta for Alzheimer's disease pathogenesis. J Neuroinflammation. 2014;11:48. doi: 10.1186/1742-2094-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J, Alzheimer Genetic Analysis Group TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, et al. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, Holtzman DM, Cirrito JR, Colonna M. TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie RH, Freeman M, Hyman BT. Expression of the macrophage scavenger receptor, a multifunctional lipoprotein receptor, in microglia associated with senile plaques in Alzheimer's disease. Am J Pathol. 1996;148:399–403. [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson K, El Khoury J. Microglial scavenger receptors and their roles in the pathogenesis of Alzheimer's disease. Int J Alzheimers Dis. 2012;2012:489456. doi: 10.1155/2012/489456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Khoury J, Hickman SE, Thomas CA, Cao L, Silverstein SC, Loike JD. Scavenger receptor-mediated adhesion of microglia to beta-amyloid fibrils. Nature. 1996;382:716–719. doi: 10.1038/382716a0. [DOI] [PubMed] [Google Scholar]
- 19.Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer's disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–565. doi: 10.1016/s0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 20.Chung H, Brazil MI, Irizarry MC, Hyman BT, Maxfield FR. Uptake of fibrillar beta-amyloid by microglia isolated from MSR-A (type I and type II) knockout mice. Neuroreport. 2001;12:1151–1154. doi: 10.1097/00001756-200105080-00020. [DOI] [PubMed] [Google Scholar]
- 21.Frenkel D, Wilkinson K, Zhao L, Hickman SE, Means TK, Puckett L, Farfara D, Kingery ND, Weiner HL, El Khoury J. Scara1 deficiency impairs clearance of soluble amyloid-beta by mononuclear phagocytes and accelerates Alzheimer's-like disease progression. Nat Commun. 2013;4:2030. doi: 10.1038/ncomms3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandoval K, Umbaugh D, House A. Crider A, Witt K. Somatostatin Receptor Subtype-4 Regulates mRNA Expression of Amyloid-Beta Degrading Enzymes and Microglia Mediators of Phagocytosis in Brains of 3xTg-AD Mice. Neurochem Res. 2019;44:2670–2680. doi: 10.1007/s11064-019-02890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, Takashima Y, Kawabe Y, Cynshi O, Wada Y, Honda M, Kurihara H, Aburatani H, Doi T, Matsumoto A, Azuma S, Noda T, Toyoda Y, Itakura H, Yazaki Y, Horiuchi S, Takahashi K, Kruijt JK, van Berkel TJC, Steinbrecher UP, Ishibashi S, Maeda N, Gordon S, Kodama T. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 24.Zhu C, Schwarz P, Abakumova I, Aguzzi A. Unaltered Prion Pathogenesis in a Mouse Model of High-Fat Diet-Induced Insulin Resistance. PLoS One. 2015;10:e0144983. doi: 10.1371/journal.pone.0144983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frontzek K, Pfammatter M, Sorce S, Senatore A, Schwarz P, Moos R, Frauenknecht K, Hornemann S, Aguzzi A. Neurotoxic Antibodies against the Prion Protein Do Not Trigger Prion Replication. PLoS One. 2016;11:e0163601. doi: 10.1371/journal.pone.0163601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell MD, Lopez-Gonzalez R, Lawson L, Hughes D, Fraser I, Gordon S, Perry VH. Upregulation of the macrophage scavenger receptor in response to different forms of injury in the CNS. J Neurocytol. 1994;23:605–613. doi: 10.1007/BF01191555. [DOI] [PubMed] [Google Scholar]
- 27.Grewal RP, Yoshida T, Finch CE, Morgan TE. Scavenger receptor mRNAs in rat brain microglia are induced by kainic acid lesioning and by cytokines. Neuroreport. 1997;8:1077–1081. doi: 10.1097/00001756-199703240-00003. [DOI] [PubMed] [Google Scholar]
- 28.Honda M, Akiyama H, Yamada Y, Kondo H, Kawabe Y, Takeya M, Takahashi K, Suzuki H, Doi T, Sakamoto A, Ookawara S, Mato M, Gough PJ, Greaves DR, Gordon S, Kodama T, Matsushita M. Immunohistochemical evidence for a macrophage scavenger receptor in Mato cells and reactive microglia of ischemia and Alzheimer’s disease. Biochem Biophys Res Commun. 1998;245:734–740. doi: 10.1006/bbrc.1998.8120. [DOI] [PubMed] [Google Scholar]
- 29.Bornemann KD, Wiederhold KH, Pauli C, Ermini F, Stalder M, Schnell L, Sommer B, Jucker M, Staufenbiel M. Abeta-induced inflammatory processes in microglia cells of APP23 transgenic mice. Am J Pathol. 2001;158:63–73. doi: 10.1016/s0002-9440(10)63945-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorce S, Nuvolone M, Russo G, Chincisan A, Heinzer D, Avar M, Pfammatter M, Schwarz P, Delic M, Muller M, et al. Genome-wide transcriptomics identifies an early preclinical signature of prion infection. PLoS Pathog. 2020;16:e1008653. doi: 10.1371/journal.ppat.1008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheckel C, Imeri M, Schwarz P, Aguzzi A (2020) Ribosomal profiling during prion disease uncovers progressive translational derangement in glia but not in neurons. Elife 9:e62911 [DOI] [PMC free article] [PubMed]
- 32.Bueler H, Raeber A, Sailer A, Fischer M, Aguzzi A, Weissmann C. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 34.Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Curr Opin Immunol. 2002;14:123–128. doi: 10.1016/s0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- 35.Areschoug T, Gordon S. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell Microbiol. 2009;11:1160–1169. doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 36.Canton J, Neculai D, Grinstein S. Scavenger receptors in homeostasis and immunity. Nat Rev Immunol. 2013;13:621–634. doi: 10.1038/nri3515. [DOI] [PubMed] [Google Scholar]
- 37.Aguzzi A, Barres BA, Bennett ML. Microglia: scapegoat, saboteur, or something else? Science. 2013;339:156–161. doi: 10.1126/science.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 39.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, de Pooter T, Mattheijssens M, van den Broeck M, Cuijt I, Vennekens K', de Deyn PP, Kumar-Singh S, van Broeckhoven C. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 40.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, de Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, George-Hyslop PS, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sierksma A, Lu A, Mancuso R, Fattorelli N, Thrupp N, Salta E, Zoco J, Blum D, Buee L, De Strooper B, et al. Novel Alzheimer risk genes determine the microglia response to amyloid-beta but not to TAU pathology. EMBO Mol Med. 2020;12:e10606. doi: 10.15252/emmm.201910606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartels T, De Schepper S, Hong S. Microglia modulate neurodegeneration in Alzheimer's and Parkinson's diseases. Science. 2020;370:66–69. doi: 10.1126/science.abb8587. [DOI] [PubMed] [Google Scholar]
- 44.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 45.Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, 3rd, Lafaille JJ, Hempstead BL, Littman DR, Gan WB. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 47.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 48.Husemann J, Loike JD, Anankov R, Febbraio M, Silverstein SC. Scavenger receptors in neurobiology and neuropathology: their role on microglia and other cells of the nervous system. Glia. 2002;40:195–205. doi: 10.1002/glia.10148. [DOI] [PubMed] [Google Scholar]
- 49.Cheng C, Hu Z, Cao L, Peng C, He Y. The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin. J Biol Chem. 2019;294:18881–18897. doi: 10.1074/jbc.RA119.010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer's disease mice. J Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Bernhardi R, Cornejo F, Parada GE, Eugenin J. Role of TGFbeta signaling in the pathogenesis of Alzheimer's disease. Front Cell Neurosci. 2015;9:426. doi: 10.3389/fncel.2015.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blots using commercial anti-Msr1 antibodies that could not detect specific Msr1 band on brains collected from wild type mice. Msr1-/- mouse brains were used as negative control. (PNG 512 kb)
RNA-Seq data of Msr1 (A) and microglia markers (Aif1, Cd68 and Itgax) (B-D) expression in hippocampi at different time points after prion inoculation. (n=3, except n=2 at 20wpi for NBH, *P<0.05; ***P<0.001). (E) Ribosomal profiling of Msr1 expression in CX3CR1-positive microglia (n=3, except n=2 at terminal stage, ****P<0.0001). Abbreviations: Msr1, macrophage scavenger receptor 1 gene; Aif1, Ionized calcium binding adaptor molecule 1 gene; Cd68, cluster of differentiation 68 gene; Itgax, integrin subunit alpha x gene; RML6, Rocky Mountain Laboratories scrapie strain, passage 6; NBH, noninfectious brain homogenates. (PNG 451 kb)
Full images of the cropped Western blots in Figure 1C (A-B), 2C (C), 3B (D-E) and 3D (F-G). (PNG 802 kb)
Data Availability Statement
The authors confirm that the data and material supporting the findings of this study are available within the article and its supplementary materials.
Not applicable.