Figure 3:
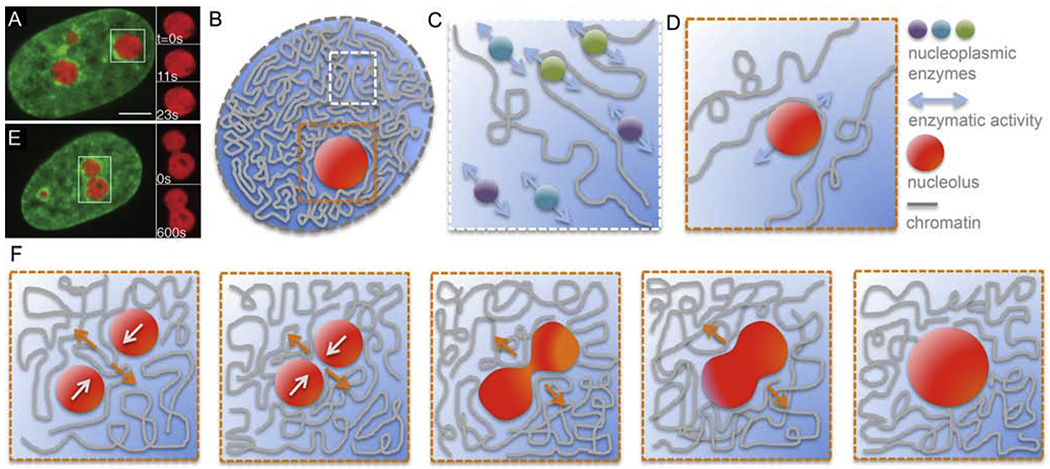
Role of nucleoplasm in chromatin dynamics. (A) Micrograph of a HeLa cell nucleus with fluorescently labeled chromatin (green, H2B-GFP) and nucleoli (red, NPM-mApple). Inset shows an enlarged boxed nucleolus at times t = 0, 11 and 23 s [54]. (B) Cartoon of a cell nucleus containing chromatin (grey), subnucleolar bodies such as nucleolus (red), and nucleoplasm (blue). White box highlights a region containing chromatin and nucleoplasm, orange box shows a region around a nucleolus. (C) Enlarged view of the white boxed region from (B) showing a schematics of free nucleoplasmic enzymes interacting hydrodynamically with each other leading to a nematic (parallel) alignment of their force dipoles. This could lead to a generation of local nucleoplasmic flows contributing to local chromatin dynamics. (D) Enlarged view of the orange boxed region from (B) showing a schematics of a moving nucleolus sterically interacting with chromatin fiber and thus leading to local rearrangement and motion of chromatin. This might occur also for nucleoplasmic enzymes or other subnuclear bodies. (E) Micrograph of a HeLa cell nucleus with fluorescently labeled chromatin (green, H2B-GFP) and two coalescing nucleoli (red, NPM-mApple). Inset shows an enlarged view of the boxed nucleolar coalescenc at times t = 0 and 11 s [54]. (F) Schematics of nucleolar coalescence leading to large-scale chromatin reorganization and dynamics inside the nucleus. Orange and white arrows indicate the local motion of chromatin and nucleoli, respectively. (A) and (E) adapted from [54]. Scale bar, 5 μm.
