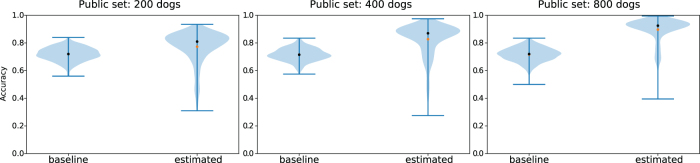
FIG. 4.
Accuracy at reconstruction of genomes x0 using EM estimation and a noisy estimate , as compared with a natural baseline that always predicts the most common variant at each SNP locus. We use this as a baseline, because without any additional information about and , the most accurate prediction of the dog's genotype would be to predict the most common variant at each locus. Here, we define accuracy as the proportion of SNPs that are correctly identified in the dog that was found in the second GWAS study, but not the first. Each distribution is constructed from 500 experimental test points, in which we (1) took 10 random splits of the full dog dataset, assigning dogs to either the public or private dataset; (2) for each split, we tested the reconstruction 50 times, each time adding a different randomly sampled dog to the second GWAS study. The private dataset always has 1000 individuals; the public test dataset is of increasing size, improving performance. EM, expectation–maximization.