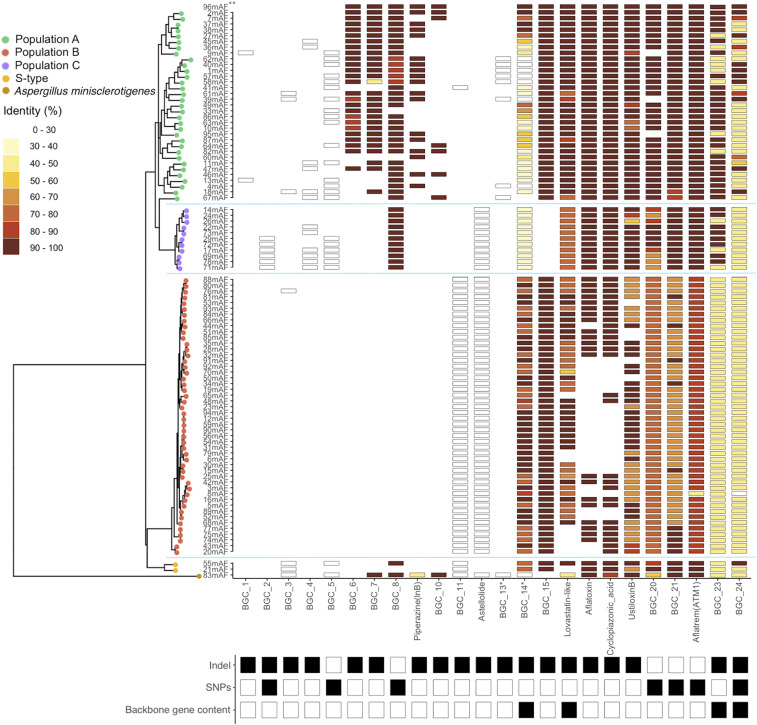
Fig. 1.
Heatmap showing BGCs with confirmed population-specific patterns among three genetically isolated L-type A. flavus populations, A (green), B (orange), and C (purple) identified previously (27) and confirmed in this study (SI Appendix, Fig. S1). In addition to 92 L-type A. flavus isolates, two S-strains (55mAF and 21mAF, Bottom, yellow) and one Aspergillus minisclerotigenes isolate (83mAF, Bottom, brown—formerly identified as Aspergillus texensis [Materials and Methods]) were included for context. The presence of a BGC, as indicated with a black-bordered box, was determined through network analysis of antiSMASH results that uses total gene content of BGCs or by previous studies (Materials and Methods). Absence of a BGC is indicated by blank spaces. When a BGC is not present in the reference genome, all corresponding boxes are white (BGCs 1 to 5 and 11 to 13). The protein sequence identity of backbone genes (core biosynthetic genes) was determined relative to the NRRL3357 reference genome (**a high-quality assembly [96mAF] was used as the reference and was compared with an assembly [2mAF—which is located immediately below 96mAF in the figure] that resulted from methodology used on all other isolates [Materials and Methods]; the high-quality assembly was not included in phylogeny). Genetic relationships among isolates are depicted on the left side using a maximum likelihood tree based on genome-wide SNPs that were thinned to be at least 3 kb apart. Population structure was determined using discriminant analysis of principle components. Population-specific variation was categorized below the heatmap as being caused by insertion/deletion (indel) or differences in protein sequence identity of backbone genes (SNPs). A third type of variation was identified when the previous types of variation occurred differentially across multiple backbone genes in a single cluster (backbone gene content). The protein sequence identity and number of backbone genes, including those found by reciprocal best-hit BLAST but not incorporated into a BGC by antiSMASH, are indicated for all 92 BGCs analyzed in SI Appendix, Fig. S2 (reference SI Appendix, SI Text for validation and incorporation of reciprocal best-hit genes). *These BGCs are physically overlapping but were distinguished by antiSMASH based on differences in gene content (see main text).