Table 1.
PISA analysis of intra- and interdimer interfaces in wt PKA R1α
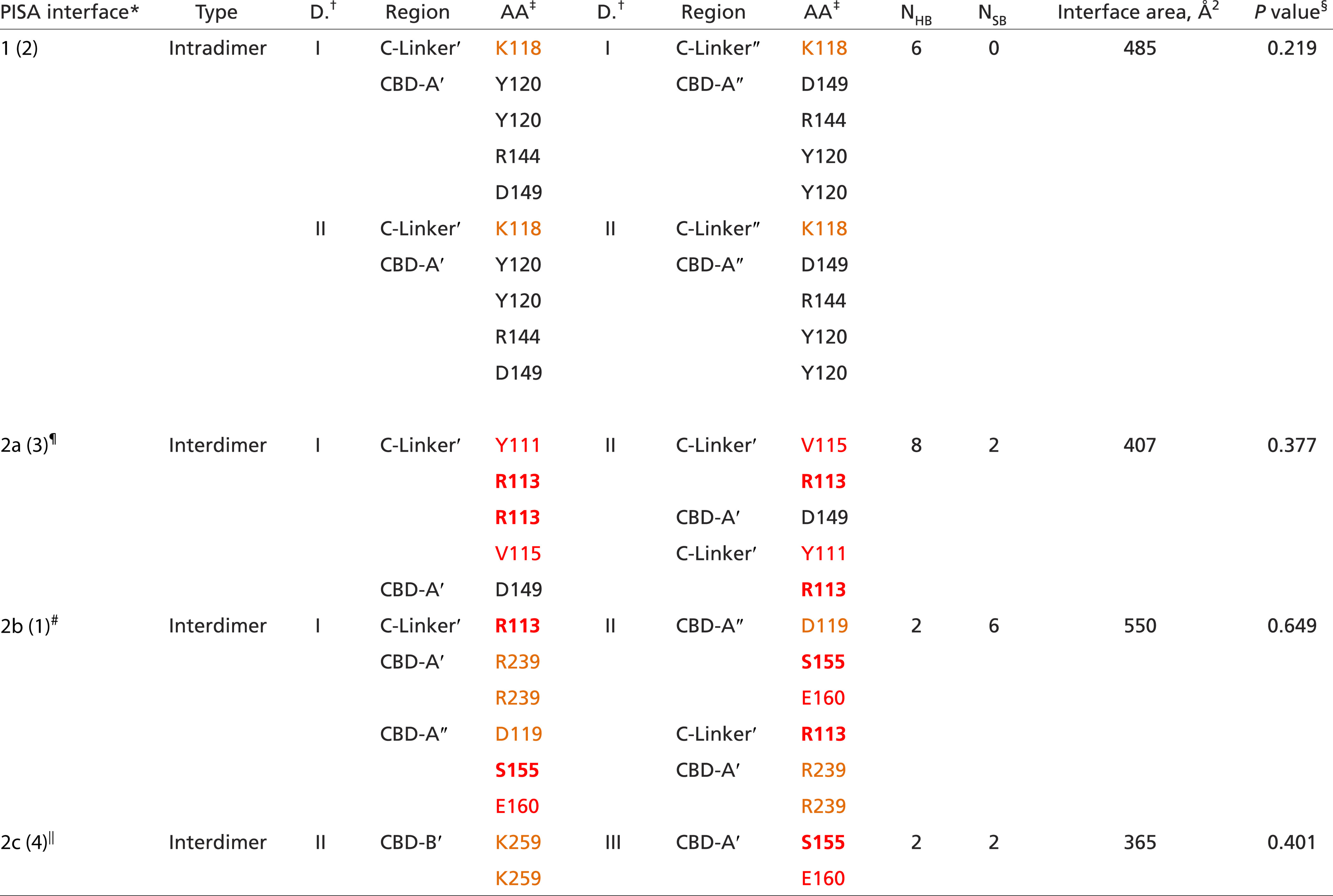 |
The 4MX3 structure was used. The numbers in parentheses indicate the interface numbering used in the PISA analysis of structure 4MX3 (68). Only interfaces including at least one hydrogen bond and/or salt bridge are included.
Dimer involved in the interface.
Residues involved in interfacial hydrogen bonds and/or salt bridges. Red means they are identified by sequence-based AGGRESCAN as belonging to an aggregation “hot spots.” Orange means they are within two residues in the primary sequence from an AGGRESCAN aggregation hot spots. Residues in bold are also predicted by AmylPred2 as consensus sites for amyloid propensity.
A value of P > 0.5 indicates that the interface is likely to reflect primarily crystal packing, while a value of P < 0.5 points to interfaces with hydrophobicity higher than average, suggesting an interaction-specific surface.
This interface includes all residues in the 108–117 segment as well as S145 and D149. A similar interface is found with P value of 0.292 in structure 1RL3 of PKA R1α 91–379 with cGMP bound to CBD-A and apo CBD-B solved at 2.70-Å resolution. Even lower values of P < 0.25 are associated with a similar interface in structures 1NE4 and 1NE6 of PKA R1α 91–379 with Rp- or Sp-cAMPS bound at both CBDs, respectively, solved at 2.40- and 2.30-Å resolution.
This interface includes also residues 108, 111, 137, 138, 140–142, 145, 146, 236, 240, 243, 244, 271, 273, 344 from C-linker′ (I) and CBD-A′ (I), and 117, 120, 121, 125, 154, 156–159, 177, 183, 214–216, 218, 220 from C-linker″ (II) and CBD-A″ (II) and the symmetrical counterparts.
This interface could not be found in the A211D structure. Further details are available in SI Appendix, Fig. S5.
