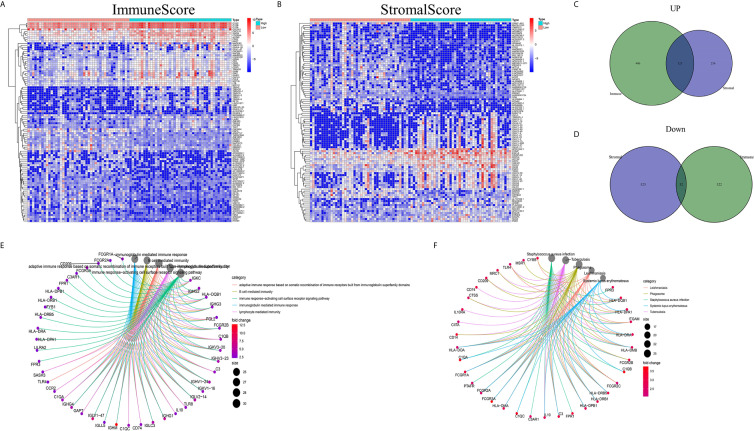
Figure 2.
DEG heatmaps, Venn diagrams, and GO and KEGG enrichment analyses. (A) Heatmaps for DEGs identified by comparing the high-score and low-score ImmuneScore groups. The gene name is shown in the row, and the sample ID (column name) is not shown. The Wilcoxon rank-sum test was used to identify differentially expressed genes, and the threshold of significance was set at q=0.05 and a fold change after log2 conversion >1. (B) DEG heatmaps, similar to (A), produced by analysis related to the StromalScore. (C, D) The Venn diagrams displaying the shared up- or downregulated DEGs identified by the analyses using the ImmuneScore and StromalScore, with a filtering threshold of significance of q<0.05 and a fold change after log2 conversion >1. (E, F) GO and KEGG enrichment analyses were carried out for the 155 DEGs, with p and q < 0.05.