Fig. 2. MYB3R4 transiently localizes in the nucleus to activate the expression of mitotic cell cycle genes.
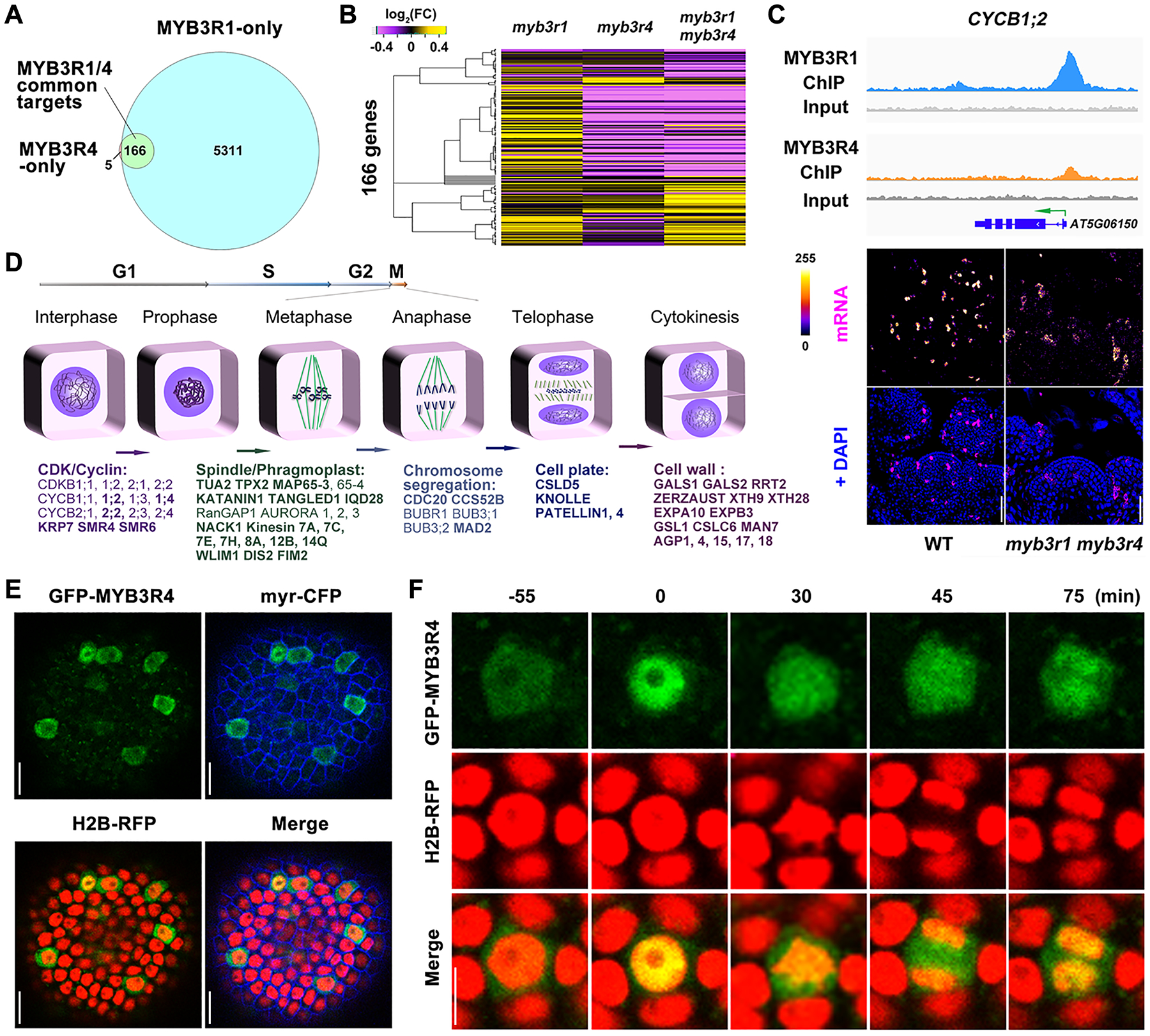
(A) Venn diagram showing the number of gene regions bound by MYB3R1 and MYB3R4 as detected by ChIP-seq.
(B) Hierarchical clustering of 166 MYB3R1 and MYB3R4 common target genes based on their relative expression levels, as detected by RNA-seq in shoot apices of single and double mutants compared to wild type. Yellow indicates increased expression and purple indicates decreased expression.
(C) MYB3R1 and MYB3R4 bind to target gene promoters and activate their expression in dividing cells. The top panels show genome browser tracks of MYB3R1 and MYB3R4 ChIP-seq coverage at a representative target gene CYCB1;2. The bottom panels show CYCB1;2 expression in wild-type and myb3r1 myb3r4 SAMs as revealed by RNA fluorescence in situ hybridisation (FISH). FISH signals are displayed using the Fire lookup table of Fiji (ImageJ) software. Scale bars, 50 μm.
(D) Classification of MYB3R1 and MYB3R4 targets based on their molecular functions. MYB3R1- and MYB3R4-regulated genes are involved in all key steps of mitotic progression. The common targets are shown in bold text.
(E) Nucleo-cytoplasmic shuttling of MYB3R4. Shown are SAM cells expressing GFP-MYB3R4 (green) together with a plasma membrane marker myr-CFP (blue) and a nuclear reporter H2B-RFP (red). Scale bars, 20 μm.
(F) The dynamic localization of GFP-MYB3R4 protein during cell division. MYB3R4 shows rapid translocalization from the cytoplasm to the nucleus at the onset of mitosis (time zero). Scale bar, 5 μm.
