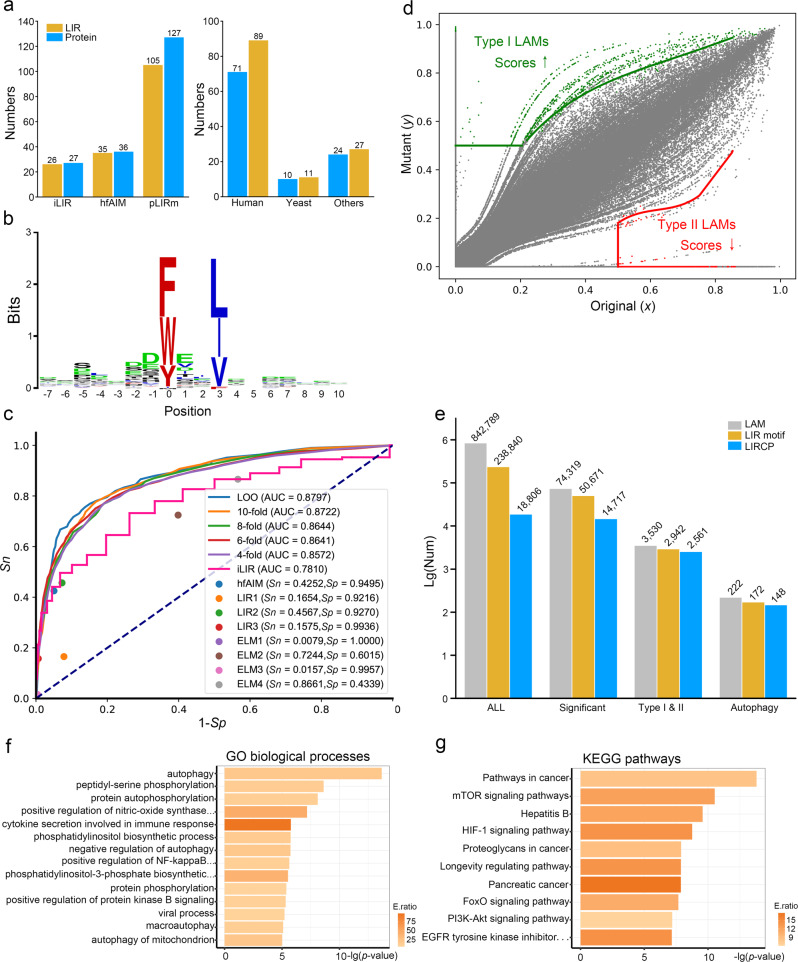
Fig. 2. Computational prioritization of highly potential LAM-containing LIRCPs.
a A comparison of known LIR motifs and corresponding proteins collected by iLIR19, hfAIM20, and pLIRm, as well as the distribution of our collected data in H. sapiens and S. cerevisiae and other species (Supplementary Data 1). b A sequence logo of known LIR motifs was generated by WebLogo (http://weblogo.berkeley.edu/logo.cgi)76. c A comparison of pLIRm to other methods, including iLIR19, hfAIM20, three LIR motifs (WXXL, [ADEFGLPRSK][DEGMSTV][WFY][DEILQTV][ADEFHIKLMPSTV][ILV], and [DE][DEST][WFY][DELIV]x[ILV])5,19,77, and four ELM motifs ([EDST].{0,2}[WFY]..P, [EDST].{0,2}[WFY][^RKPG][^PG][ILV], [EDST].{0,2}LVV, and [EDST].{0,2}[WFY]..[ILVFY])21. d The model-based algorithm pLAM for predicting Type I and Type II LAMs that potentially increase and decrease the binding affinity of cLIR motifs to LC3, respectively. e The distribution of numbers of potential LAMs, LIR motifs and LIRCPs reserved in each step of pLAM. f, g The GO- and KEGG-based enrichment analyses of finally reserved LAM-containing LIRCPs.