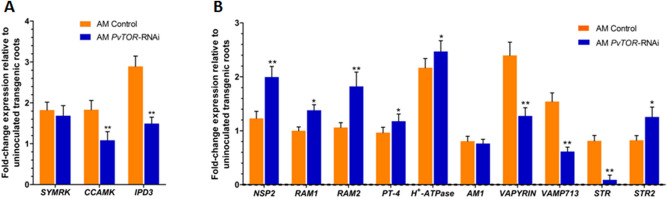
Figure 8.
Expression of P. vulgaris AM regulatory genes in R. irregularis-inoculated transgenic roots. (A) RT-qPCR analysis of the transcript levels of CSSP genes and AM fungi induced TFs genes in control and PvTOR-RNAi transgenic roots at 1 wpi. (B) RT-qPCR analysis of the transcript levels of mycorrhizal-induced genes in control and PvTOR-RNAi transgenic roots at 1 wpi. (A & B) Quantitative RT-PCR was performed on cDNA of root RNA samples. The statistical significance of differences between mycorrhized roots of control and PvTOR-RNAi plants was determined using an unpaired two-tailed Student’s t-test (*P < 0.05; **P < 0.01). Error bars refer to the SE of the mean of three biological replicates (n > 9). CSSP, common symbiotic signaling pathway; AM, arbuscular mycorrhiza; TFs, transcription factors; wpi, week post inoculation.