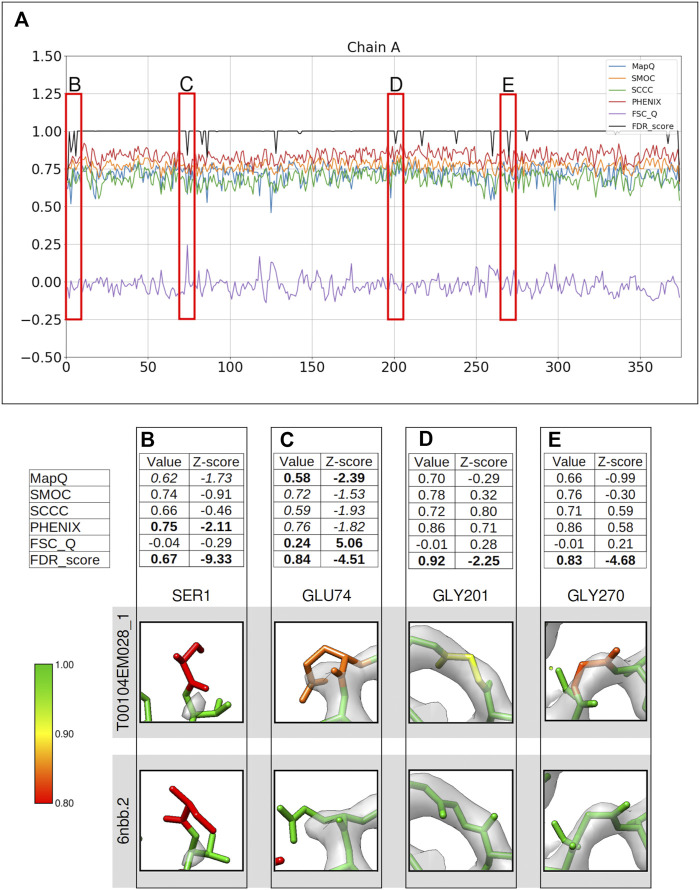
FIGURE 2.
Comparison of metrics for the atomic model of alcohol dehydrogenase (color bar shows the correspondence with the FDR scores assigned, residues in red have FDR scores around 0.8 or worse, yellow around 0.9, and green around 1.0). The metrics are calculated only for the backbone atoms. (A) Per-residue plot of the scores from MapQ, SMOC, SCCC, PHENIX, FSC-Q, and FDR backbone score for the chain A of the atomic model T0104EM028_1 from the EMDB model; the red boxes highlight the residues selected for detailed analysis in the panels below. For Ser1 (B), Glu 74 (C), Gly 201 (D), and Gly 270 (E), the panel shows a table with values of scores obtained with each metric and corresponding Z-scores; the residue fit in the target map (EMD-0406, grey) displayed at the recommended contour level and rendered in UCSF Chimera; and the residue fit in map as modeled in the reference (PDB ID: 6nbb.2).