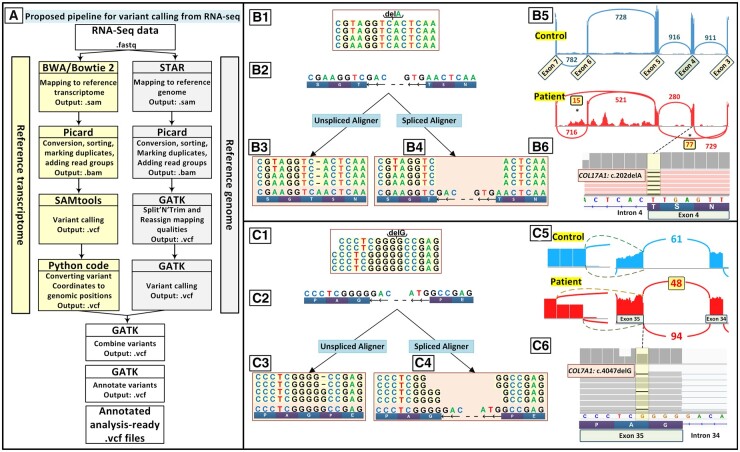
Fig. 2.
Parallel use of a reference genome and a reference transcriptome improves variant calling from RNA-seq data for Mendelian disorders. (A) Workflow of our proposed pipeline for variant calling from RNA-seq data with 2 parallel processes for mapping of RNA-seq data to the reference genome and reference transcriptome. (B) Mapping to the reference genome detected the mutation c.5422C>T in EXPH5 (data not shown) in a patient with EB with 2 homozygous mutations but not a deletion mutation c.202delA in COL17A1, which was detected by mapping to the reference transcriptome. (B.1) Schematic representation of RNA sequenced reads for the homozygous mutation c.202delA in COL17A1. (B.2) Reference genome corresponding to the location of sequenced reads with a negative-strand orientation. (B.3) RNA sequenced reads when mapped to the reference genome by a splice aligner tool such as STAR. (B.4) RNA sequence reads were mapped with the reference transcriptome by unspliced aligners tools. (B.5) A sashimi plot revealed that the c.202delA mutation at the end of exon 4 leads to complex aberrant splicing of COL17A1 pre-mRNA, including skipping of exons 4 and 6 in 77 and 15 sequencing reads, respectively. (B.6) Screenshot of the genomic sequence visualized by IGV demonstrating the homozygous mutation COL17A1: c.202delA. (C.1) Schematic representation of sequenced reads harboring the heterozygous deletion of c.4047delG in COL7A1 by RNA sequencing in a patient affected by DEB with compound heterozygous deletion mutations of c.3840delC and c.4047delG. (C.2) Reference genome corresponding to the location of sequenced reads with a negative-strand orientation. (C.3) Mapped reads to the reference genome show a lower number of reads for the first 2 nucleotides of an exon and extra reads for the last nucleotide of the adjacent exon and first nucleotide at the beginning of the intron. (C.4) Sequences mapped to the reference transcriptome clearly show the heterozygous deletion. (C.5) A sashimi plot revealed that c.4047delG in exon 35 of COL7A1 leads to aberrant splicing. (C.6) Screenshot of IGV demonstrating the heterozygous mutation COL7A1:4047delG.