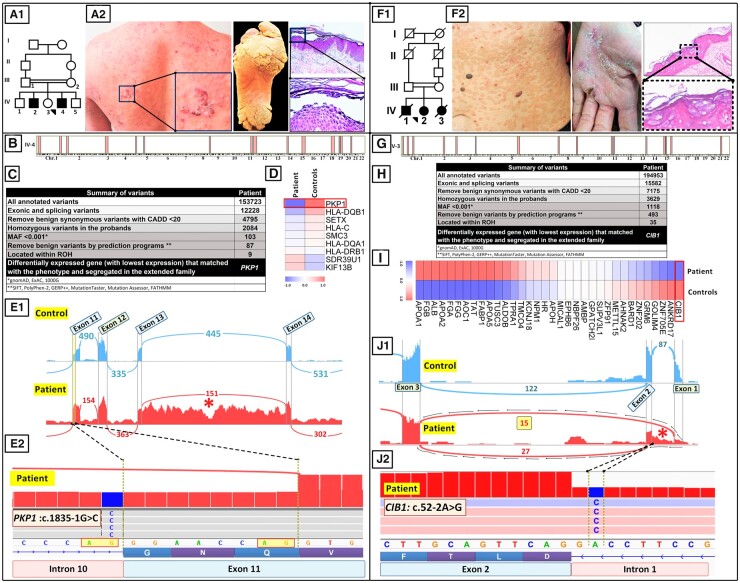
Fig. 3.
RNA-seq as a first-tier method of mutation detection in skin diseases. (A–F) Case 1. Family pedigree (A.1) and clinical features (A.2) in a patient with a skin fragility disorder with hyperkeratosis. (B) HM based on RNA-seq data revealed 15 ROHs in the proband. (C) Bioinformatics filtering steps of annotated variants that were used to narrow the candidate gene down to PKP1, which has been associated previously with a skin fragility syndrome. (D) A heat map of extracted DEGs revealed that the expression of PKP1 was most downregulated among the 9 genes coinciding with the ROHs. (E.1) A Sashimi plot revealed that this mutation resulted in aberrant splicing of exon 11 and retention of intron 13 sequences (asterisk). (E.2) IGV visualization of the genomic sequence revealed a homozygous mutation, PKP1:c.1835-1G>C affecting the last nucleotide in intron 10. (F–J) Case 2. Family pedigree (F.1) and clinical features (F.2) and histopathology of a patient with EV. (G) RNA-seq–based HM revealed 17 ROHs in the proband. (H) Stepwise bioinformatics filtering and coalignment of the putative mutant genes with ROHs identified a pathogenic variant in CIB1. (I) CIB1 was the most downregulated gene among the 35 variants coinciding with ROHs. (J.1, J.2) Variant calling from RNA-seq data identified a homozygous mutation, CIB1:c.52-2A>G. The Sashimi plot revealed complex aberrant splicing, including partial intron 1 retention and exon 2 skipping (asterisk).