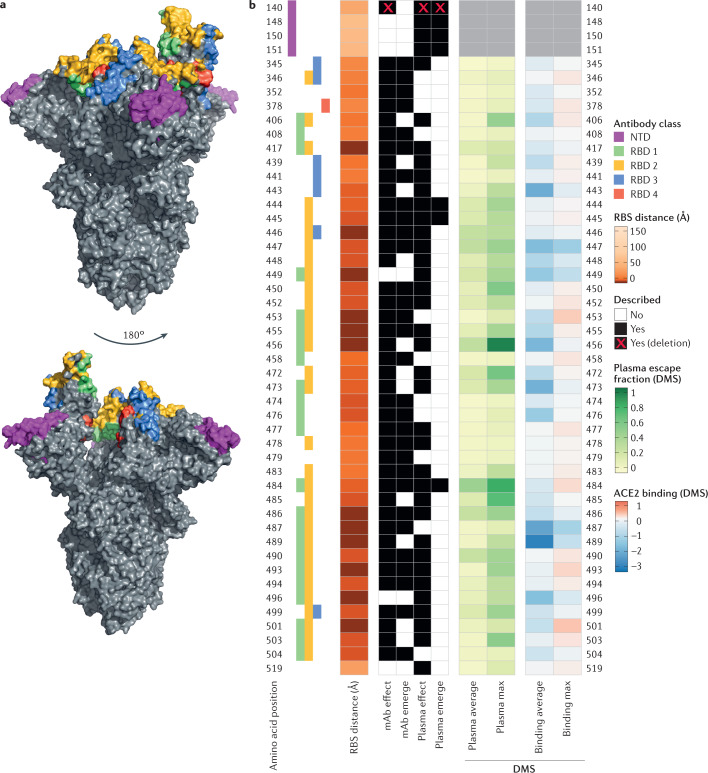
Fig. 1. Neutralizing antibody classes defined by structural analyses and properties of spike protein residues.
a | Amino acid residues of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein are coloured according to the class of the antibody that binds to an epitope. Receptor-binding domain (RBD) antibody classes 1–4 (ref.31) are shown: green for class 1 (ACE2-blocking antibodies that bind the spike protein in the open conformation), yellow for class 2 (ACE2-blocking antibodies that bind the RBD in both the open conformation and the closed conformations), blue for class 3 (antibodies that do not block ACE2 and bind the RBD in both the open conformation and the closed conformations) and red for class 4 (neutralizing antibodies that bind outside the ACE2 site and only in the open conformation). When residues belong to epitopes of multiple classes, priority colouring is given to antibodies that block ACE2 and bind the closed spike protein. The amino-terminal domain (NTD) supersite30 is coloured in magenta. b | Aligned heat maps showing properties of amino acid residues where substitutions affect binding by antibodies in polyclonal human blood plasma or emerge as antibody escape mutations. The distance in angstroms to the ACE2-contacting residues that form the receptor-binding site (RBS) is shown in shades of orange; each residue is classified as having evidence for mutations affecting neutralization by either monoclonal antibodies (mAbs)40,43,47,48 or polyclonal antibodies in plasma from previously infected individuals (convalescent)39–41,43,48 or vaccinated individuals59 (‘mAb effect’ and ‘plasma effect’, respectively). A subset of these residues has mutations described as emerging upon exposure (co-incubation) to mAbs40,47,48 or plasma40,41 in laboratory experiments (‘mAb emerge’ and ‘plasma emerge’, respectively). When an observation includes a deletion, this is indicated by a red cross. Shades of green depict the results of deep mutational scanning (DMS) experiments where yeast cells expressing RBD mutants are incubated with multiple samples of human convalescent plasma39. The escape fraction (that is, a quantitative measure of the extent to which a mutation reduced polyclonal antibody binding) averaged across all amino acid substitutions at a residue (‘plasma average’) and the maximally resistant substitution (‘plasma max’) are indicated. DMS data on ACE2-binding affinity19 are shown in shades of red or blue representing higher or lower ACE2 affinity, respectively. The mean change in binding affinity averaged across all mutations at each site (‘binding average’) and alternatively the maximally binding mutant (‘binding max’) is shown. Scores represent binding constants (Δlog10 KD) relative to the wild-type reference amino acid.