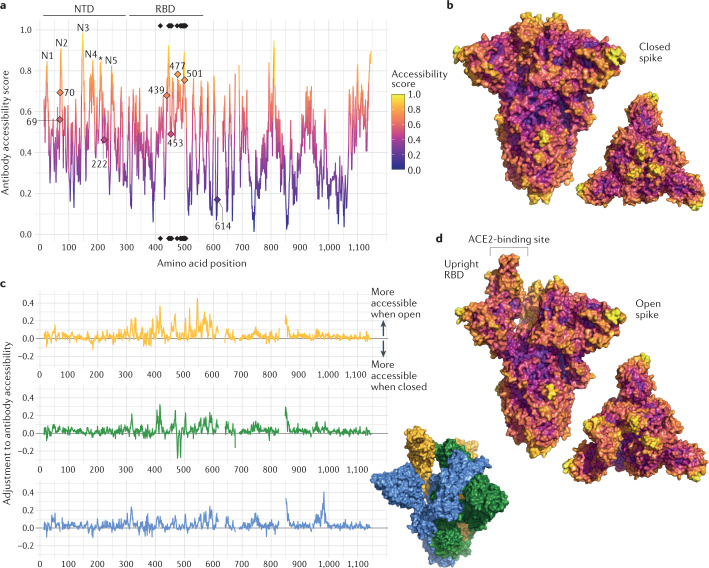
Fig. 2. Structure-based analysis of conformational epitopes on the spike protein.
a | Structure-based antibody accessibility scores for each spike protein ectodomain residue in the closed form were calculated with BEpro49. Black diamonds at the top and bottom of the plot indicate the positions of ACE2-contacting residues. Accessible amino-terminal domain (NTD) loops N1–N5 are labelled, and a loop falling between these is indicated with an asterisk. b | Two surface colour representations of antibody accessibility scores for the spike protein in the closed conformation according to the colour scheme in part a: a trimer axis vertical view (left) and an orthogonal top-down view along this axis (right). c | The extent to which each spike residue becomes more or less accessible when the spike protein is in its open form is shown. For each spike monomer (upright receptor-binding domain (RBD) (yellow), closed RBD clockwise adjacent (green) and closed RBD anticlockwise adjacent (blue)), the difference relative to the score calculated for the closed form (shown in part a) is shown. d | Two surface colour representations of antibody accessibility scores for the spike protein in the open conformation with a single monomer with an upright RBD are shown: a trimer axis vertical view (left) and an orthogonal top-down view along this axis (right).