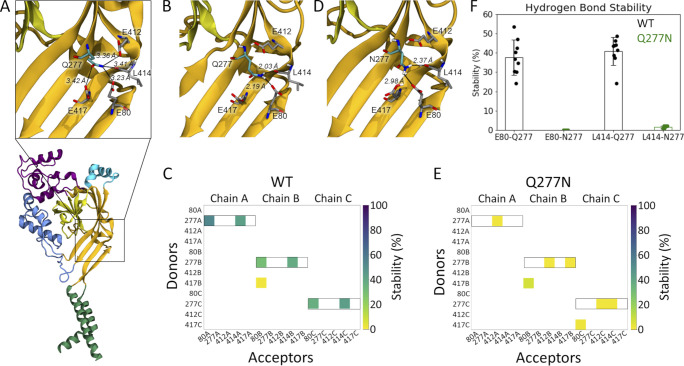
Figure 4.
Gln277 links Leu414 and Glu80 via hydrogen bond network. (A) A single subunit of cASIC1 in the desensitized state, illustrating residues within potential hydrogen bonding distance of Gln277 (inset). Colors as in Fig. 1 A. (B) Snapshot from a WT simulation illustrating the hydrogen bond network with Gln277 in the center, hydrogen bonding to L414 and E80. The snapshot was taken at 8.6 ns. (C) Hydrogen bond analysis for a representative repeat (100 ns) of WT with E80 deprotonated and E412 and E417 protonated. All hydrogen bonds formed between donors and acceptors of the side chains of E80, Q277, E412, and E417 are considered, as well as hydrogen bonds in which the backbone oxygen atom of L414 participates as an acceptor. Acceptors are listed horizontally, donors vertically. The colored squares illustrate that a given hydrogen bond is present for part of the 100 ns of simulation, following the color bar given to the right. Hydrogen bonds in which Q277 participates as a donor are highlighted by black boxes. (D) Snapshot from a Q277N simulation illustrating that the inserted Asn residue is too short to form the same hydrogen bond network as Gln277. The snapshot was taken at 19.2 ns. (E) Hydrogen bond analysis as in D, but for the Q277N mutant. (F) Average stability (bars) of the E80-Q277 and the L414-Q277 hydrogen bonds in the WT (black) and Q277N simulations (green; WT: E80-Q277: 38 ± 9%; L414-Q277: 41 ± 7%; Q277N: E80-N277: 0.02 ± 0.01%; L414-N277: 1.6 ± 0.6%. The nine data points (three chains × three repeats) are illustrated as points, and the error bar depicts SD.