Figure 2.
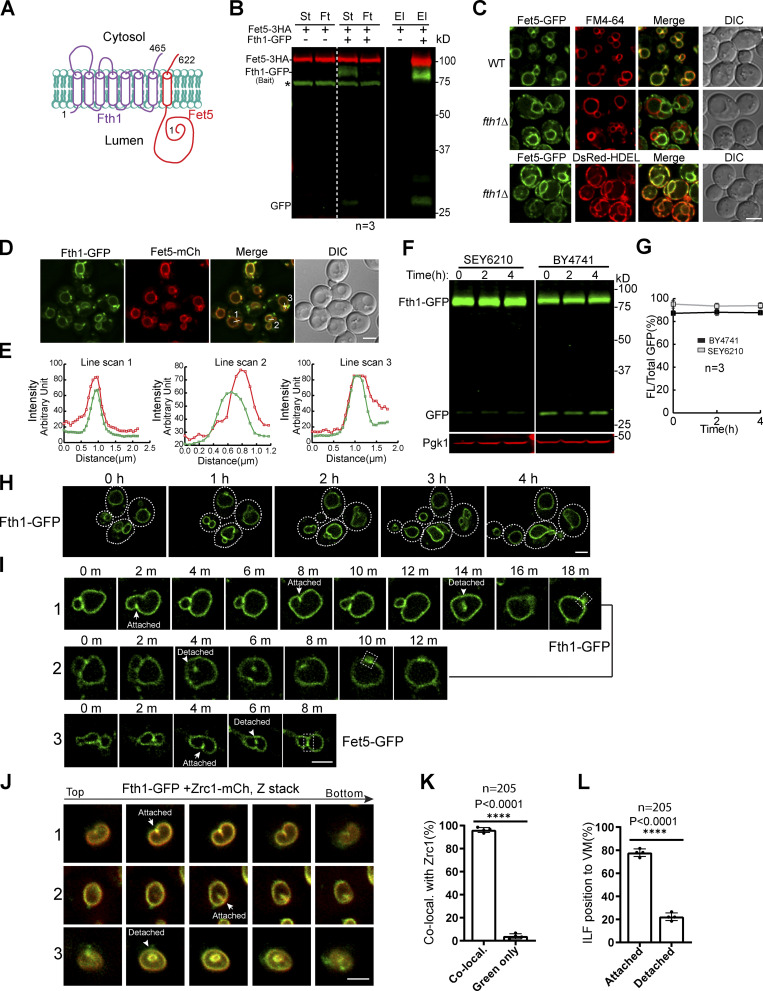
Fth1 forms a stable complex with Fet5 and is not constitutively degraded.(A) Membrane topology of the Fth1–Fet5 complex. (B) Coimmunoprecipitation showing the interaction between Fth1 and Fet5. El, elution; Ft, flow through; St, starting material. Asterisk indicates the nonspecific band. (C) The complex formation is essential for Fet5 to exit the ER. FM4-64 and DsRed-HDEL highlight VM and ER, respectively. (D) Fth1-GFP colocalizes with Fet5-mCherry at the boundary membrane of adjacent vacuoles. (E) Line scans to show the colocalization in D. (F) Fth1-GFP is not constitutively degraded in either SEY6210 or BY4741 background. (G) Quantification (±SD, n = 3) of protein levels in F. (H) Time-lapse imaging to show no increase of lumenal GFP during normal vacuole fusion and fission processes. Dashed lines indicate the periphery of yeast cells. (I) Time-lapse imaging to show the formation of ILFs and their fusion back to the VM. (J and K) Images (J) and quantification (K) showing the colocalization of Fth1-GFP with Zrc1-mCherry on ILFs. Each data point in K represents a biological repeat. A total of 205 ILFs were counted. (L) Distribution of the ILFs relative to the VM. DIC, differential interference contrast. Scale bars, 3 µm.