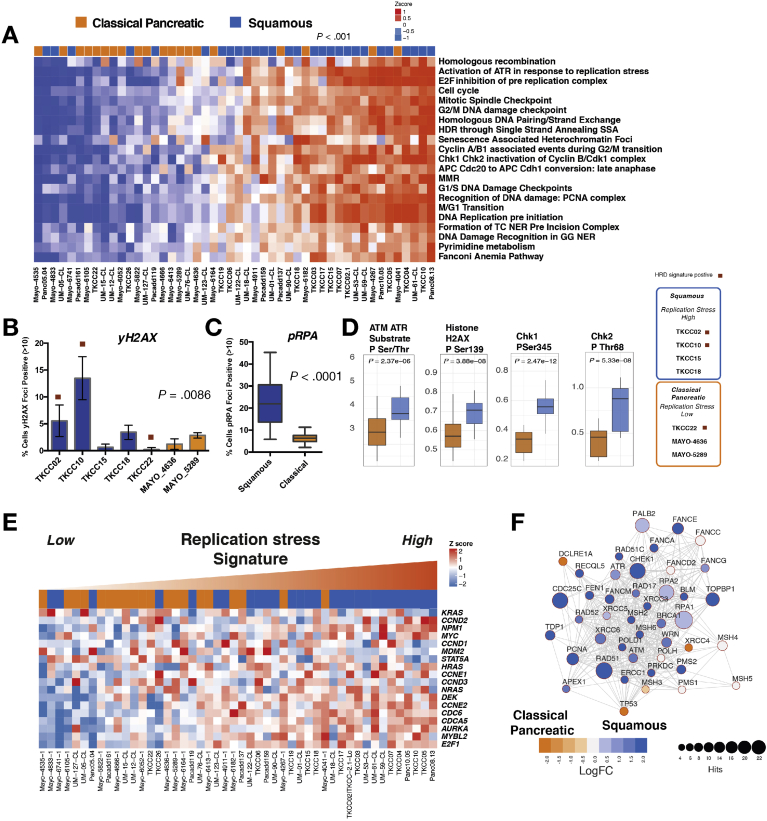
Figure 3.
Replication stress in PDCLs of PC. (A) Heatmap of pathways and molecular processes (GO terms) involved in DNA maintenance and cell cycle regulation activated in replication stress and DNA damage response. PDCLs are ranked from right to left based on the decreasing novel transcriptomic signature score of replication stress, and molecular subtype is indicated in the top bar showing the association between activation of replication stress and the squamous subtype (P < .001, chi-square test, low vs high). (B, C) Immunofluorescent quantifications of (B) γH2AX and (C) pRPA at normal conditions are elevated in the squamous (blue) but not the classical pancreatic (orange) PDCLs. (D) Proteomic analysis using RPPA of a panel of PDCLs showed that replication stress response proteins are differentially activated in the squamous subtype. (E) Heatmap showing oncogene expression in PDCLs ranked from right to left by replication stress signature. Squamous PDCLs are enriched for oncogene activation and replication stress. (F) siRNA screening showing transcriptome functional interaction subnetwork, showing preferential dependencies of cell cycle control and DNA maintenance genes in the squamous subtype. Different node colors represent dependencies in different molecular subtypes, and the size of each node is relative to the number of siRNA hits. FC, fold change.