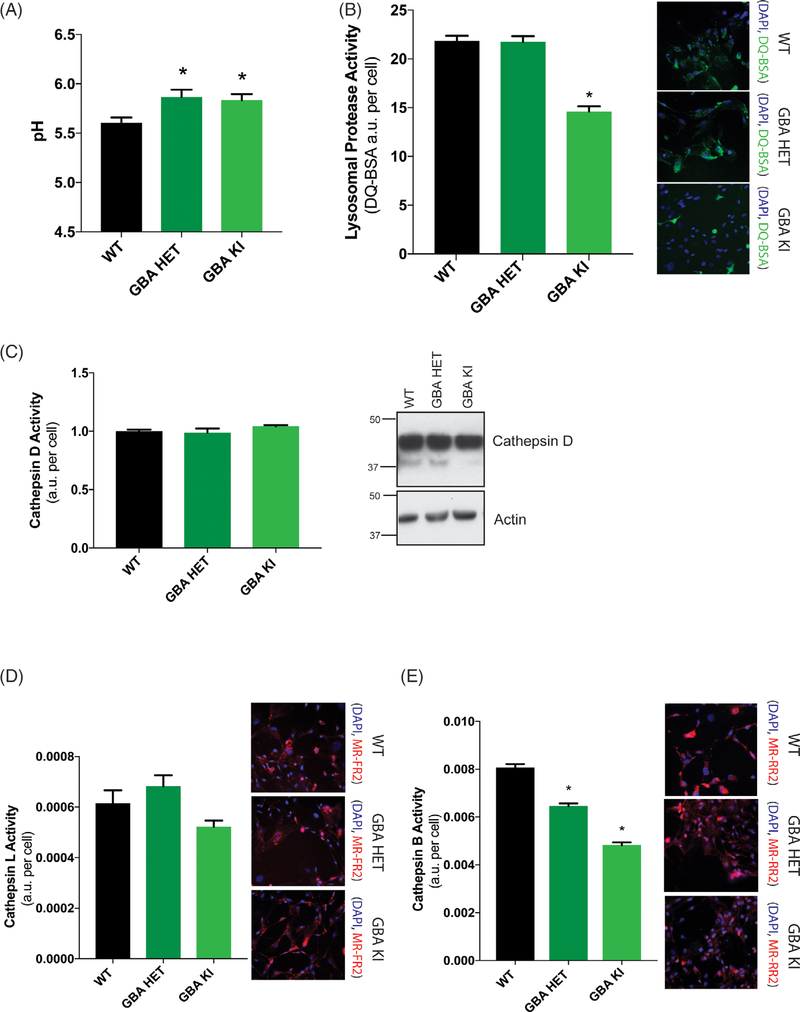
FIG. 2.
GBA1 D409V knockin mutation reduces lysosomal protease activity. (A) Determination of lysosomal pH using LysoSensor displays increased alkalinization to pH 6 in heterozygous (GBA HET) and homozygous (GBA KI) knockin astrocytes. Data are collated from 3 independent experiments with 20 wells per genotype (N = 3, F = 5.017, P = 0.0089). (B) General lysosomal protease activity, as detected by DQ-BSA cleavage (green), shows a significant reduction in homozygous (GBA HET), but not heterozygous (GBA KI), knockin astrocytes. Green fluorescence as a result of cleavage of DQ-BSA is observed in representative microscopy images (N = 3, F = 55.41, P < 0.0001). (C) Cathepsin D activity, as detected by cathepsin D activity assay and Western blot of cathepsin D protein band and (D) cathepsin L activity, as detected by cleavage of Magic-Red substrate (red), exhibit no change as a result of a GBA1 mutation (N = 3, F = 3,783, P = 0.0309). (E) Cathepsin B activity, as detected by cleavage of Magic-Red substrate (red), show a significant dose-dependent reduction in heterozygous (GBA HET) and homozygous (GBA KI) knockin astrocytes. Red fluorescence from cathepsin B/L-specific substrate cleavage is observed in representative microscopy images (N = 3, F = 161.3, P < 0.0001). In all fluorogenic plate-based activity assays, the nuclei were detected by Hoechst 33258 (blue) for normalization of the fluorescent signal. Data were collated from 3 independent experiments with 5 to 10 wells per genotype per experiment (N = 3). *P < 0.05, analysis of variance followed by Tukey’s post hoc test.