Figure 3.
HDX-MS mapping of RNA binding to NNTD-LKR S176D/S188D/S206D
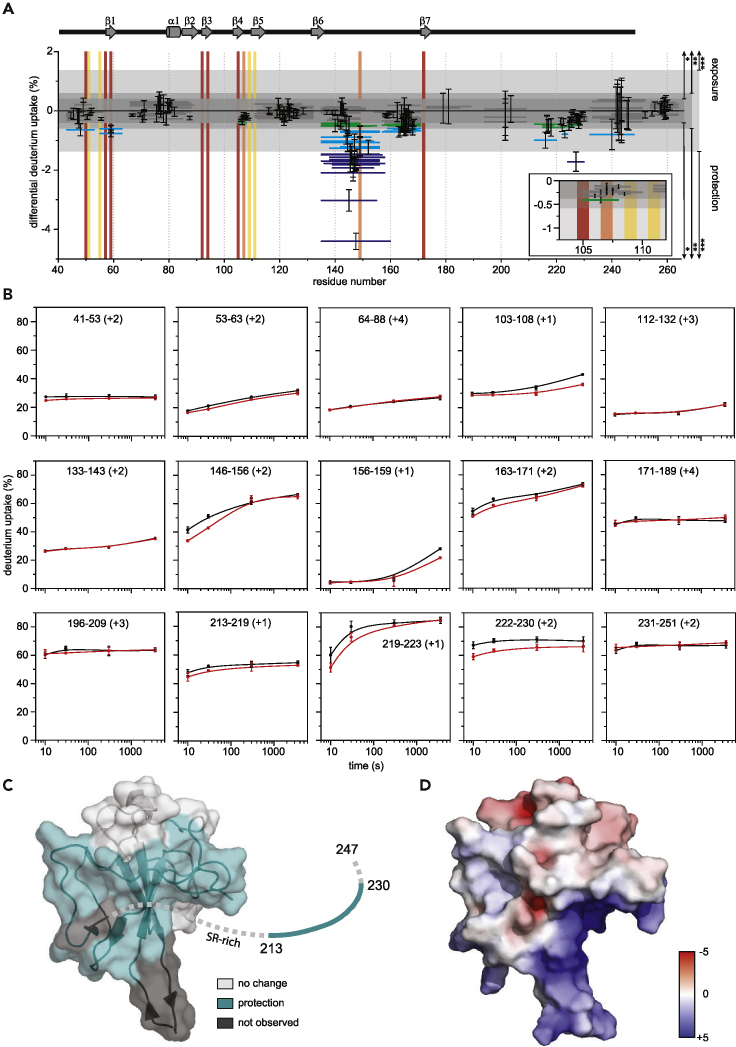
(A) Woods' plot showing cumulative differential HDX and validating differences using global significance limits. The horizontal bars depict the cumulative HDX differences between the RNA-bound and unbound NNTD-LKR S176D/S188D/S206D. Standard deviations are shown for each peptide. Peptides showing statistically significant differences are differentiated by global significance limit using the standard error of the mean and t-values for a two-tailed Student's t-distribution (∗, p < 0.1; ∗∗, p < 0.05; ∗∗∗, p < 0.01). The blue shade of the peptide bar indicates differing statistical significance (green, light blue, and dark blue, respectively); gray peptide bars depict peptides where statistically significant differences in HDX were not observed. Vertical bars show previously reported binding sites (residues reported for RNA-binding CoV2 N-protein (Dinesh et al., 2020, Ye et al., 2020), AMP-binding HCoV-OC43 (Lin et al., 2014; Ye et al., 2020), and for both are shown in red, yellow, and orange, respectively). Secondary structure of SARS-CoV-2 NNTD (PDB 6M3M) is shown above.
(B) Representative kinetic plots showing peptide level HDX as a function of exchange time (unbound, black; bound to RNA, red).
(C) Sites of protection measured by HDX mapped on the NNTD structure (PDB 6M3M). Statistically significant HDX protection, regions of no difference in HDX, and regions where lacking proteolytic coverage results in no data are shown in teal, light gray, and dark gray, respectively. Those residues unresolved in the structure are shown as a dashed line, with the exception of those reporting a statistically significant difference in teal.
(D) Electrostatic potential calculated with APBS mapped on to the NNTD structure (PDB 6M3M) shows a major positive charge groove. Red and blue represent negative and positive electrostatic potential. The color scale is in kTe−1 units. See also Figure S4 and Mendeley data set (https://doi.org/10.17632/sv8r6phkzt.1).