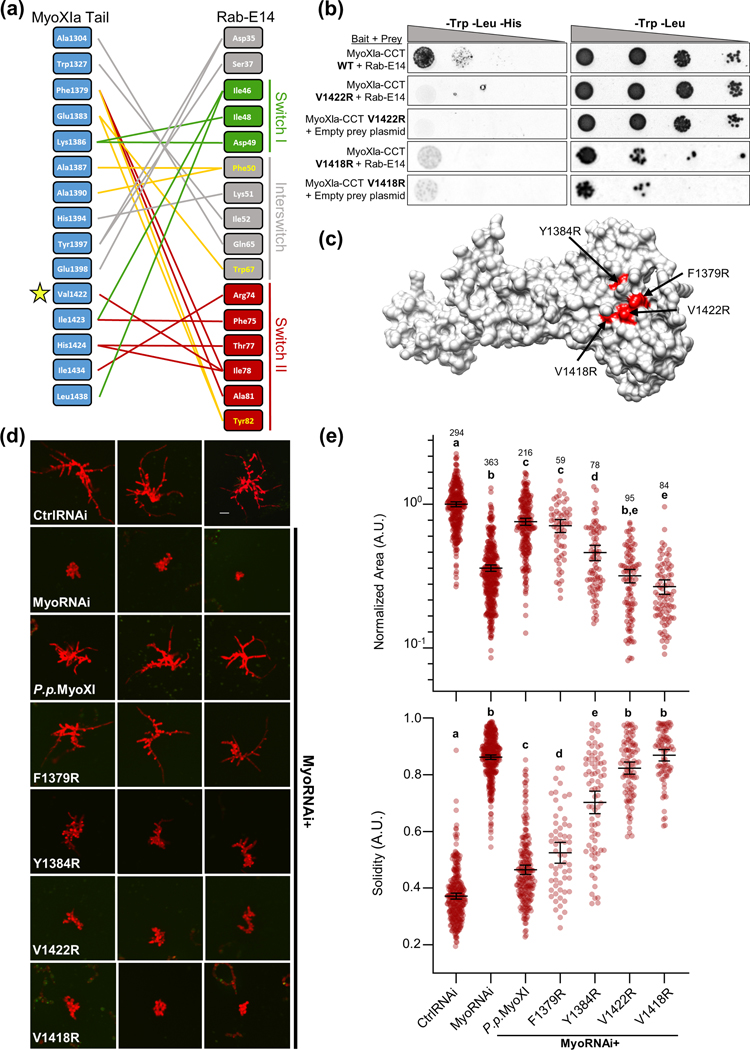
Figure 4. Structure-guided mutagenesis of predicted Myosin XI-CBD:Rab-E14 binding interface reveals polarized growth mutants in Physcomitrella patens.
(A) Contact map of modeled myosin XI:Rab-E14 (−17.8 kcal mol-1) determined by template-based docking algorithm PRISM (Tuncbag et al., 2011). Hydrophobic triad residues and their putative contacts colored in yellow. Colors of lines representing contacts reflect the domain within Rab-E14 that the contact belongs. Green=Switch I, Red= Switch II, Grey=Interswitch, Yellow=Hydrophobic triad.
(B) Directed Y2H with the P.p. myoXI-CCT WT bait fragment from (Fig. 3) and bait fragments containing mutations in the myosin XI cargo-binding domain that are required for polarized growth and predicted to interact with Rab-E14. All yeast strains were grown on the same SD plates, but rearranged for clarity.
(C) Homology model of P.p. myosin XI-tail is shown, with the candidate interface residues predicted to mediate Rab-E interactions in red.
(D) Representative 1-week old RNAi plants—all plants except ‘CtrlRNAi’ are silencing expression of both endogenous myosin XIs. All plants except ‘MyoRNAi’ were co-transformed with the myosin XI silencing construct and another construct expressing either WT myosin XI or mutant myosin XI. Bar = 100 μm.
(E) Quantification of the morphological parameters of solidity and area extracted from images in (d). Error bars indicate standard error of the mean. Shared letters above the bars show those experimental groups that cannot be statistically distinguished. Statistical significance was determined by a one-way ANOVA-Tukey (P<0.01). Numbers above the letters indicate number of plants analyzed per condition.