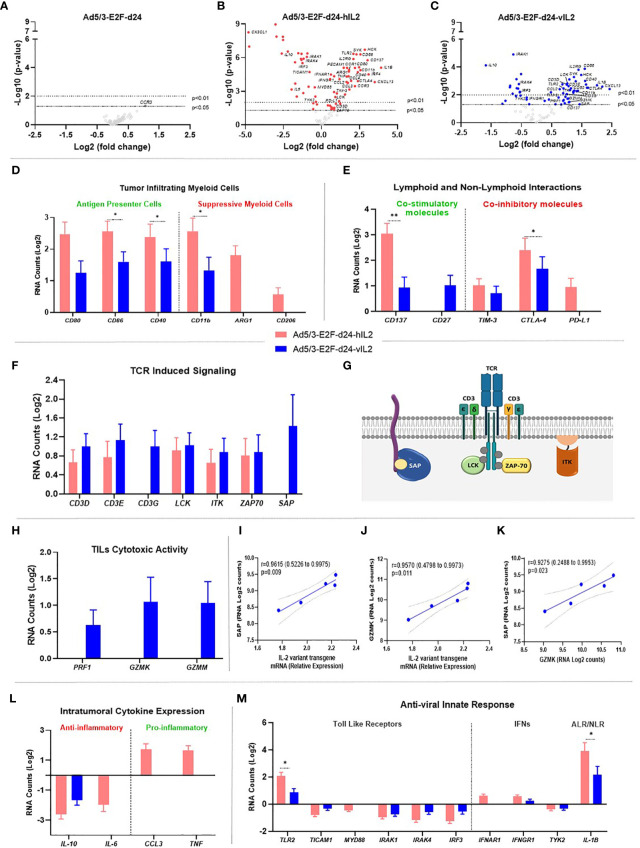
Figure 5.
Molecular gene expression analysis in the virus treated tumors. (A–C) Volcano plot graphs showing each gene’s -log10(p-value) and log2 fold change with expression change upon (A) Ad5/3-E2F-d24, (B) Ad5/3-E2F-d24-hIL2, and (C) Ad5/3-E2F-d24-vIL2 treatments. Statistically significant genes fall at the top of the plot above the horizontal lines, and differentially expressed genes fall to left and right sides on the X axis. The dashed horizontal lines represent the significance levels thresholds: p<0.05 or p<0.01. (D) Log2 mRNA expression of genes associated to antigen presenting cells (left side) and to suppressive myeloid cells (right side). (E) Lymphoid and non-lymphoid interactions: mRNA (Log2) expression study on co-stimulatory (left side) and co-inhibitory molecules (right side). (F) Log2 mRNA expression of genes associated to TCR induced signaling. (G) Graphical illustration of TCR signaling involved proteins on the surface of lymphocyte cells. (H) Log2 mRNA expression of TIL cytotoxicity genes. (I) Correlation between SAP and vIL-2 transgene expression levels. (J) Correlation between GZMK and vIL-2 transgene levels. (K) Correlation between SAP and GZMK expression levels. (L) Intratumoral expression of anti-inflammatory cytokines (left side) and pro-inflammatory cytokines (right side) as mRNA (Log2) levels. (M) Anti-viral innate immune response related to TLRs (left side), IFNs (middle), and inflammasome signaling via AIM2-like receptors (ALRs) and NOD-like receptors (NLRs) (right side) as mRNA (Log2) expression. All data is presented as mean + SEM. *p < 0.05, **p < 0.01.