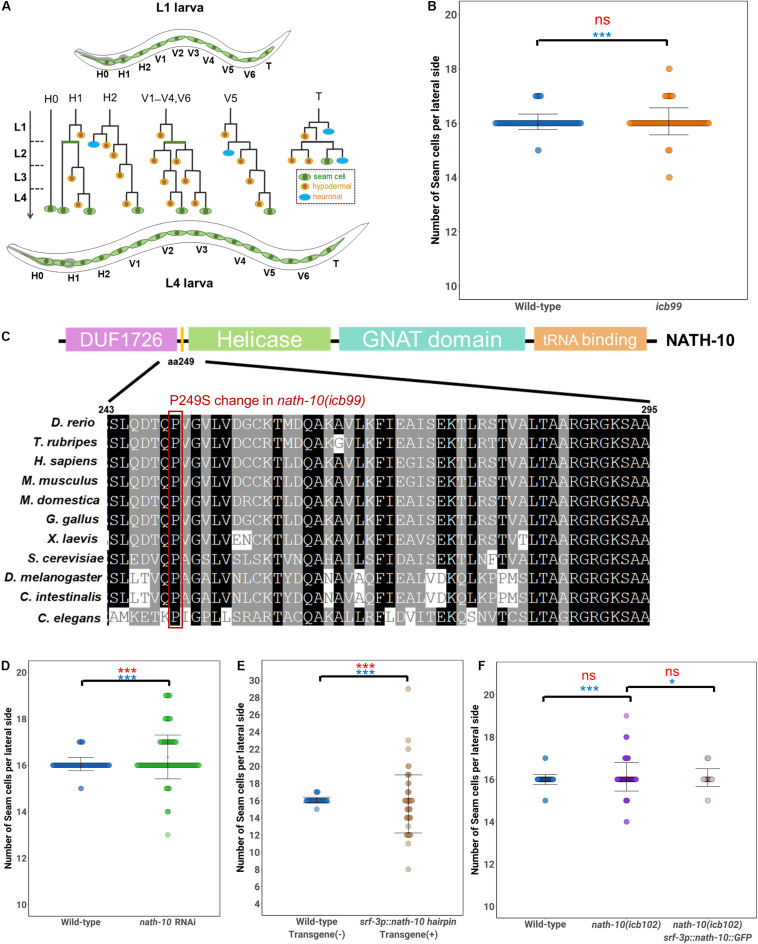
FIGURE 1.
Seam cell number robustness is disrupted by nath-10 loss-of-function. (A) schematic displaying seam cell lineage from L1 to L4, seam cells are labeled green, hypodermal cells are labeled yellow and neurons are labeled blue. (B) Mutants carrying the icb99 mutation display variable seam cell number (SD = 0.28, n = 120 for wild type JR667 animals, SD = 0.50, n = 120 for animals carrying the icb99 mutations. Red ns indicates non-significant difference to the mean by t-test (p > 0.05). Blue stars indicate statistically significant difference to the variance by Levene’s median test (***p < 0.001). (C) Schematic showing protein domains in NATH-10. The P249S change involves a proline to serine shift at amino acid 249, indicated by the yellow bar between the DUF1726 and Helicase domains. Alignment of this region across multiple species shows high conservation of this proline, suggesting it may be of importance to protein function. Red frame shows the conserved proline. (D) nath-10 RNAi phenocopies the icb99 mutation leading to a significant increase in variance and average seam cell number (Levene’s median test ***p < 0.001 and t-test, ***p < 0.001, n = 120 wild type, and n = 183 nath-10 RNAi). (E) Seam cell specific knockdown of nath-10 by a nath-10 hairpin RNAi also causes significant increase in seam cell number and seam cell variance in comparison to wild-type animals not carrying the hairpin transgene (t-test, ***p < 0.001, Levene’s test median ***p < 0.001). (F) A CRISPR replacement mimicking the icb99 mutation phenocopies the original EMS-derived mutant (SD = 0.77, Levene’s test ***p < 0.05). Seam cell specific overexpression of nath-10 (srf-3p:nath-10:GFP) partially rescues this phenotype (SD = 0.41, Levene’s test *p < 0.05 In (B,D–F) mean values are indicated by red dots and error bars correspond to ± Standard deviation. Statistical differences to the mean and variance are indicated above brackets for relevant comparisons in red and blue, respectively, assessed by a t-test or Levene’s median test. Significance levels (ns p > 0.05, *p < 0.05, ***p < 0.001).