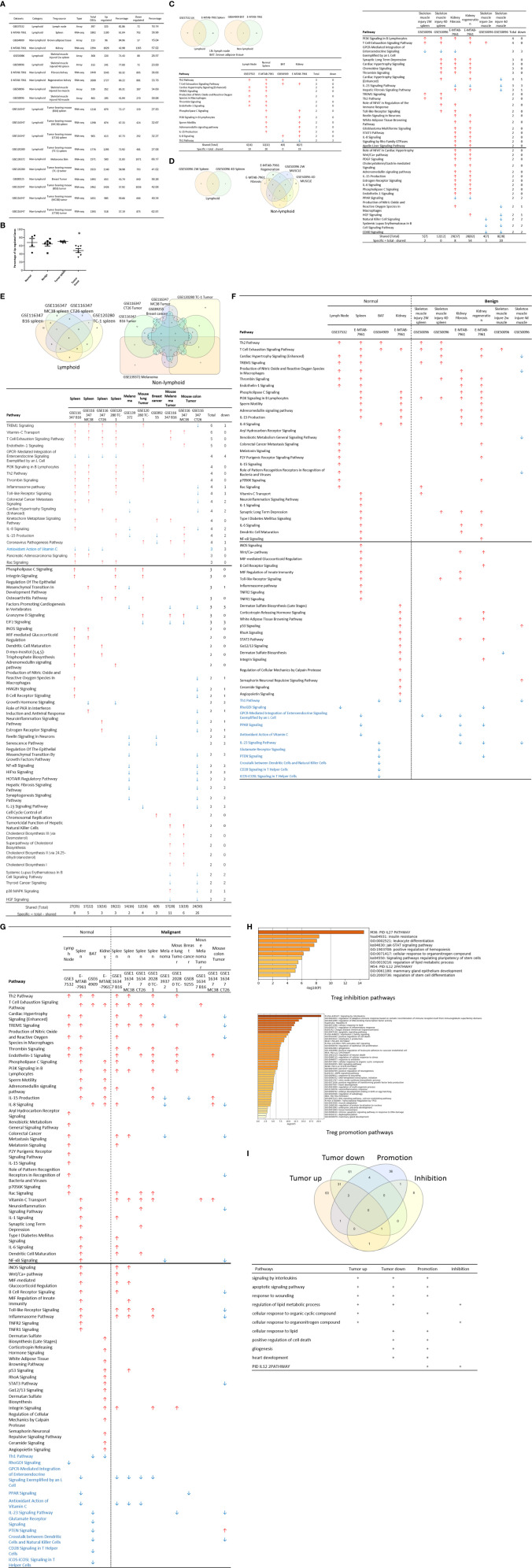
Figure 2.
(A) Comparisons of regulatory T cells (Treg) versus conventional T cells (Tconv) in tissues enrolled in Table 1 led to identification of differentially expressed genes (DEGs) in each dataset. The type of results from datasets enrolled including Array data and RNA-seq data. (Cut off: p < 0.05; |Log2FC| ≥ 1). (B) There were more suppressed DEGs of Treg in Tumor tissues than other 3 groups. The mean percentages of up-regulated genes in tumor tissue Treg groups were 48.22 ± 5.004, lower than that in normal, benign, and tumor spleen groups (67.71 ± 9.806, 64.20 ± 4.891, 70.04 ± 1.458, respectively). *Significantly changed. (C) The majority (73.8%, 59/80) of enriched pathways in normal tissue Treg groups were tissue Treg-specific in lymphoid and non-lymphoid tissues. The ingenuity Pathway Analysis (IPA, Qiagen, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) using the differentially expressed gene (DEG) from datasets enrolled in this study showed the activated (↑) and inhibited (↓) canonical pathways in normal tissue Treg groups (p < 0.05, |Z-score| >= 2). In addition, Venn diagram analysis was performed to find the commonly shared pathways in this group. The specifically enriched pathways in GSE37532 Lymph node (LN), E-MTAB-7961 spleen, and E-MTAB-7961 Kidney were shown in Supplementary Table 3A . In the normal Treg groups, there were a total of 80 pathways enriched including 64 activated and 16 inhibited. There were no dual regulated pathways in normal Treg groups. The most common shared pathway was Th2 Pathway, which was activated in 71.2% (5/7) datasets. TREM1 Signaling, Production of Nitric Oxide and Reactive Oxygen Species in Macrophages, Cardiac Hypertrophy Signaling (Enhanced), and Thrombin Signaling where activated only in Treg from lymphoid tissues. Whereas Th1 Pathway and tRNA Splicing were inhibited, and IL-8 Signaling was activated only in Treg in non-lymphoid tissues. (D) Non-lymphoid tissue Treg enriched more specific pathways than lymphoid tissue Treg in benign disease groups. (85 specific pathways in 4 non-lymphoid Treg datasets vs. 2 specific pathways in 2 lymphoid Treg datasets). IPA analysis using the DEG from datasets enrolled in this study showed the activated (↑) and inhibited (↓) canonical pathways in different benign disease tissue Treg groups (p < 0.05, |Z-score| >= 2). Venn diagram analysis was performed to find the commonly shared pathways in this groups. The specifically enriched pathways in various Treg groups were showed in Supplementary Table 3B . A total of 124 pathways were enriched in benign disease Treg groups, including 90 activated and 30 inhibited pathways. Cardiac Hypertrophy Signaling (Enhanced), Hepatic Fibrosis Signaling Pathway, Reactive Oxygen Species in Macrophages, and HGF Signaling were dual regulated pathways in non-lymphoid tissue Treg. The percentages of specifically enriched pathways in this group was 70.2% (87/124). 97.7% (85/87) specific pathways were in non-lymphoid tissue Treg datasets. The most commonly shared pathway in benign disease Treg groups were T Cell Exhaustion Signaling Pathway and PI3K Signaling in B Lymphocytes, activating in 67% (4/6) datasets. TREM1 Signaling and Th2 Pathway were only activated in spleen tissue Treg in this group. Whereas IL-23 Signaling Pathway, PPAR Signaling, Natural Killer Cell Signaling, Systemic Lupus Erythematosus In B Cell Signaling Pathway, and CD40 Signaling were inhibited in non-lymphoid tissue Treg. (E) Malignant Treg groups enriched more inhibited and dual pathways than normal and benign disease Treg groups. IPA analysis using the DEG from Treg datasets enrolled in this study showed the activated (↑) and inhibited (↓) canonical pathways in different malignant disease tissue Treg groups (p < 0.05, |Z-score| >= 2). Venn diagram analysis was performed to find the commonly shared pathways in this Treg group. The specifically enriched pathways in each dataset were showed in Supplementary Table 3C . There were a total of 128 pathways enriched in the malignant Treg groups, including 62 activated, 50 inhibited, and 16 dual regulated pathways. There were 19 specifically enriched pathways in 4 lymphoid Treg datasets (mean 4.8 per dataset), 52 specifically enriched pathways in 6 non-lymphoid Treg datasets (mean 8.7 per dataset). TREM1 Signaling and Vitamin-C Transport were the most commonly shared pathways. In addition, TREM1 Signaling was activated in all the lymphoid tissue Treg and dual regulated in the non-lymphoid tissue Treg. Vitamin-C Transport was activated both in lymphoid and non-lymphoid tissue Treg. (F) The Dataset GSE50096 Treg from skeletal muscle injured 4d enriched more down regulated pathways than normal and other benign disease tissue Treg. The pathways enriched in normal lymphoid and non-lymphoid tissue Treg were compared with benign tissue Treg. The 5 pathways all of which were activated in normal tissue Treg were suppressed in Treg from skeletal muscle injured 4d and 2w tissues. All the suppressed pathways in normal tissue Treg groups were also suppressed or non-statistically enriched in benign disease tissue Treg. (G) Malignant tissue Treg enriched more down regulated pathways than normal tissue Treg. The Pathways enriched in normal lymphoid and non-lymphoid tissue Tregs were compared with malignant tissue Treg. The 13 pathways all of which were activated in normal tissue Tregs were suppressed in at least one malignant Treg datasets, more specifically, in malignant non-lymphoid tissue Treg. However, only one of the 11 suppressed pathways in normal tissue Treg, PTEN Signaling, was activated in GSE116347 CT26 dataset. (H) Metascape analysis of 18 Treg inhibition genes and 57 Treg promotion genes (PMID:32680511) identified 11 Treg inhibition GO and pathways and 45 Treg promotion GO and pathways. Moreover, M54: PID IL12 2PATHWAY was a dual regulated pathway, which was enriched both in Treg promotion and inhibition pathways. (I) There were 4 suppressed and 1 activated Treg promotion pathways in malignant Treg datasets whereas there was only 1 activated Treg inhibition pathway in malignant Treg datasets. DEGs in malignant Treg datasets enrolled in this study were analyzed by metascape and compared with Treg promotion and inhibition pathways. Tumor Treg upregulated pathways included up-regulated DEGs in malignant Treg datasets whereas tumor downregulated pathways included down-regulated DEGs in malignant Treg datasets. The 4 downregulated Treg promotion pathways were cellular response to lipid, positive regulation of cell death gliogenesis, gliogenesis, and heart development. One up-regulated Treg inhibition pathway was cellular response to organonitrogen compound. The 61 Tumor downregulated, 31 dual, and 63 tumor upregulated pathways were summarized in Supplementary Table 4 .